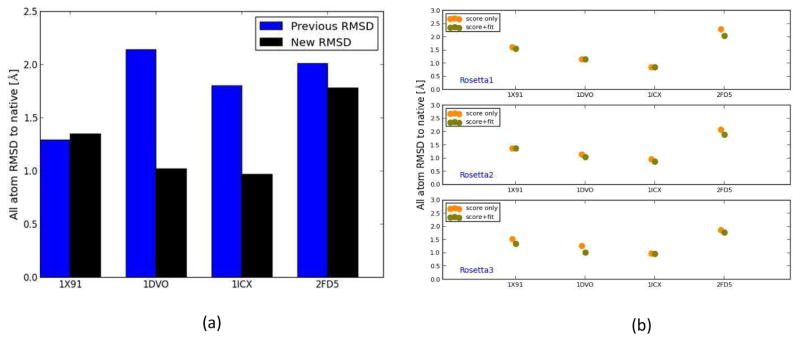
Figure 4.
(a) The RMSDs of the final refined models using iterative Rosetta-MD protocol where models were picked based on a consensus score which is a combination of the Rosetta score and the fit-to-density (black) and based on Rosetta score-only (blue). For 1DVO, 1ICX and 2FD5 the new method was able to further improve the quality of the final structures. (b) RMSD of the selected models in each Rosetta step for the soluble proteins. Orange filled circles show the RMSDs of models picked by the best score method and green filled circles show the RMSDs of models picked based on consensus score method. For 75% of the cases, the consensus score method was able to pick a better model than the score-only method.