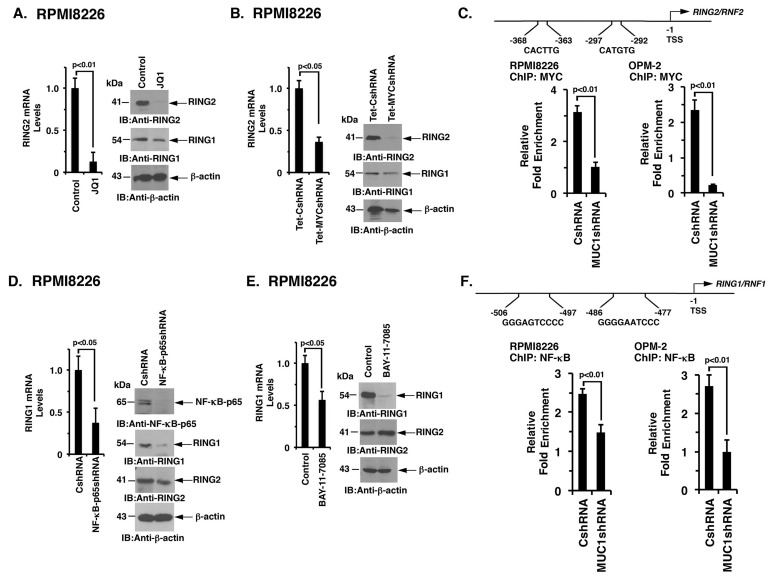
Figure 6. MUC1-C regulates RING2 and RING1 by MYC- and NF-κB p65-dependent mechanisms, respectively.
A. RPMI8226 cells treated with JQ1 or vehicle control for 48 h were analyzed for RING2 mRNA levels by qRT-PCR (left). The results (mean±SD of 3 determinations) are expressed as relative mRNA levels as compared with that obtained for the vehicle treated cells (assigned a value of 1, left). Lysates were immunoblotted with the indicated antibodies (right). B. RPMI8226/Tet-CshRNA and RPMI8226/Tet-MYCshRNA cells treated with DOX for 5 d were analyzed for RING2 mRNA levels by qRT-PCR (left). The results (mean±SD of 3 determinations) are expressed as relative mRNA levels as compared with that obtained for the Tet-CshRNA cells (assigned a value of 1). Lysates were immunoblotted with the indicated antibodies (right). C. Schema of the RING2 promoter with highlighting of putative MYC binding sites. Soluble chromatin from the RPMI8226/CshRNA and RPMI8226/MUC1shRNA (left) and OPM-2/CshRNA and OPM-2/MUC1shRNA (right) cells was precipitated with anti-MYC or a control IgG antibody. The final DNA samples were amplified by qPCR with pairs of primers (Supplementary Table S2) encompassing the MYC binding sites in the RING2 promoter. The results (mean±SE of 3 determinations) are expressed as the relative fold enrichment compared with that obtained for the IgG control (assigned a value of 1). D. RPMI8226/CshRNA and RPMI8226/NF-κBshRNA were analyzed for RING1 mRNA levels by qRT-PCR (left). The results (mean±SD of 3 determinations) are expressed as relative mRNA levels as compared with that obtained for the CshRNA cells (assigned a value of 1). Lysates were immunoblotted with the indicated antibodies (right). E. RPMI8226 cells treated with the 5 μM BAY-11-7085 or vehicle control for 24 h were assessed for RING1 levels by qRT-PCR (left). The results (mean±SD of 3 determinations) are expressed as relative mRNA levels as compared with that obtained for the vehicle treated cells (assigned a value of 1). Lysates were immunoblotted with the indicated antibodies (right). F. Schema of the RING1 promoter with highlighting of putative NF-κB p65 binding sites. Soluble chromatin from the RPMI8226/CshRNA and RPMI8226/MUC1shRNA (left) and OPM-2/CshRNA and OPM-2/MUC1shRNA (right) cells was precipitated with anti-NF-κB p65 or a control IgG antibody. The final DNA samples were amplified by qPCR with pairs of primers (Supplementary Table 2) encompassing the NF-κB p65 binding sites in the RING1 promoter. The results (mean±SE of 3 determinations) are expressed as the relative fold enrichment compared with that obtained for the IgG control (assigned a value of 1).