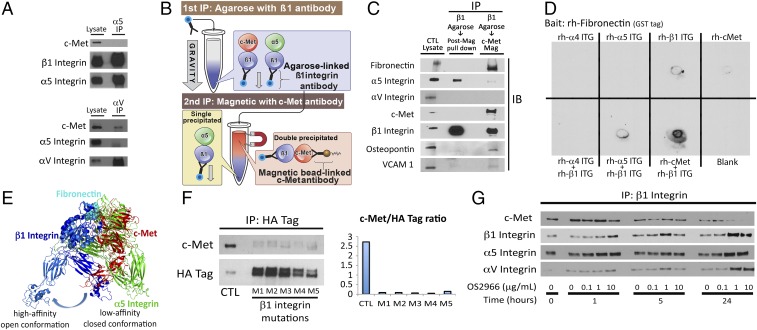
Fig. 6.
c-Met/β1 integrin complex affinities and modeling. (A) IP revealed that αV but not α5 integrin bound c-Met in U87. (B) Sequential IP schematic: isolating the c-Met/β1 complex by binding β1 via agarose beads, followed by magnetics beads binding c-Met in complex with β1. (C) Sequential IP of β1 followed by c-Met revealed that a c-Met/β1 double IP contained far more fibronectin (FN), VCAM, and osteopontin and far less α5 than a β1 integrin single IP, with αV in neither, suggesting that c-Met displaces α5 from its β1 binding site and that c-Met/β1 integrin binds FN better than α5/β1 integrin. (D) Dot blotting revealed that coincubated c-Met+β1 bound FN far better than c-Met, α4, α5, or β1 alone; α5+β1; or α4+β1. (E) PyMOL analysis defined steric constraints causing c-Met to preferentially bind β1 in its high-affinity open conformation. (F) Five amino acids in β1 integrin predicted by ALA scanning as crucial to c-Met/β1 binding were changed to alanine by mutagenesis of a β1–HA fusion protein. HA IP of HEK cells expressing these constructs revealed decreased c-Met binding to each of five mutants versus wild-type β1–HA fusion protein. (G) Treating cultured U87 cells with variable concentrations of β1 neutralizing antibody OS2966 for 1, 5, and 24 h decreased c-Met/β1 complex formation as assessed by β1 IP.