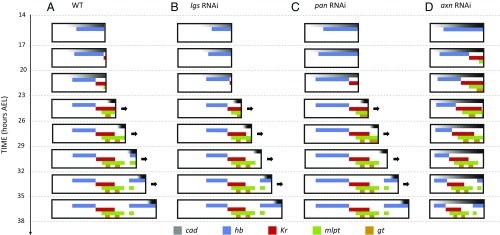
Fig. 3.
Speed regulation model recapitulates gap gene expression in Tribolium WT, lgs RNAi, pan RNAi, and axn RNAi embryos. (A) In a computer simulation, where the speed of Tribolium gap gene sequence is regulated by cad gradient (black/gray; darker corresponds to higher concentration), gap gene (hb, blue; Kr, red; mlpt, green; gt, gold) spatiotemporal dynamics were recapitulated during blastoderm and germband stages of WT Tribolium embryos (compare with Fig. 2A and Fig. S3A). (B) To simulate lgs RNAi background, cad gradient was reduced and shifted toward posterior. Accordingly, gap gene waves were slower and shifted toward posterior (compare with Fig. 2E). (C) To simulate pan RNAi background, cad gradient was reduced, stretched, and shifted toward anterior. Accordingly, gap gene waves were slower, stretched, and shifted toward anterior (compare with Fig. S8I). (D) To simulate axn RNAi background, germband elongation and cad gradient retraction were halted. Accordingly, gap gene waves continued to propagate and shrink in the germband and never stabilized (compare with Fig. S3B). See Movie S3.