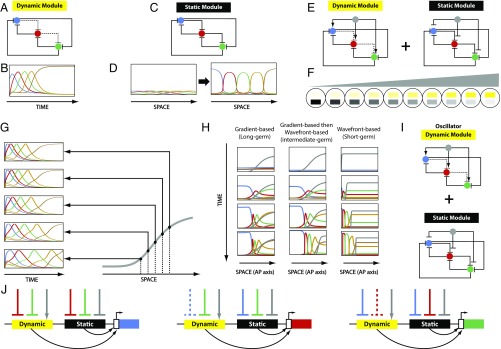
Fig. 5.
Gradual enhancer switching model. (A) A three-gene cascade (dynamic module). Solid line, strong repression; dashed line, weak repression. (B) Computer simulation of a five-gene cascade (same structure as the three-gene cascade: Every gene is strongly repressing all other genes, except only weakly repressing its immediate successor in the cascade). Shown are expression profiles of the cascade constituent genes over time. (C) A three-gene mutual exclusion gene network (static module). (D) Shown is an array of cells along a spatial axis. Active in each cell is a five-gene mutual exclusion gene network (same structure as the three-gene mutual exclusion network: Every gene is strongly repressing all others). Cells along the spatial axis are initialized with a broad and diffuse domains of the expression of five genes (Left). Over time, the diffuse domains get stronger and sharper (Right). (E) The gradual enhancer switching gene network: A speed regulator (gray) is activating all of the genes in the dynamic module, while repressing all of the genes in the static module. (F) Shown is an array of cells in which the gradual enhancer switching gene network is active. Applied to the array is a gradient of the speed regulator (gray). This results in the gradual switching from the dynamic module (yellow) to the static module (black) as we go from high to low values of the gradient. (G) Computer simulations of the gradual enhancer switching model at different positions along the spatial axis (i.e., different values of the gradient, shown in gray): The higher the concentration of the gradient, the higher the speed of sequential activation of genes. (H) Computer simulations of the gradual enhancer switching model under different conditions: nonretracting speed regulator (gradient-based, long-germ insects; Left), retracting speed regulator (wavefront-based, short-germ insects; Right), and nonretracting then retracting speed regulator (gradient-based then wavefront-based, intermediate-germ insects; Middle). See Movie S5. (I) The gradual enhancer switching model with an oscillator as a dynamic module. The model induces oscillatory kinematic waves of gene expression, observed in vertebrates and insects (Movie S7). (J) Encoding dynamic and static modules by separate enhancers ensures modularity, since each module requires different regulatory logic. Shown is the regulatory wiring required for each gene in the combined dynamic+static gene network in E. Solid lines, strong interactions; dashed lines, weak interactions.