Fig. 5.
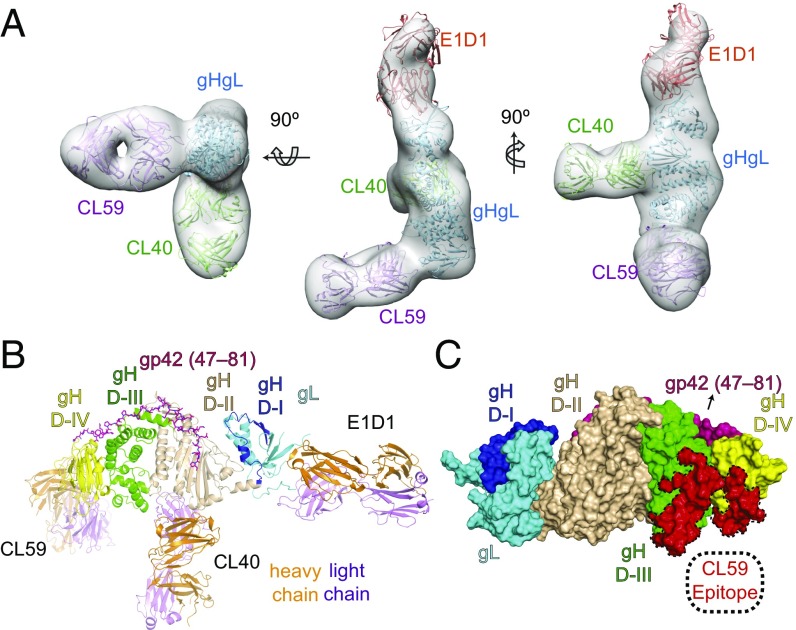
CL59 binds to an epitope spanning gH domains D-III and D-IV. (A) 3D negative-stain EM model of three Fabs (CL59, CL40, and E1D1) bound to gHgL. The EM envelope (gray) is shown with fitted crystal structures of gHgL (blue) and three Fabs represented as CL59 Fab (purple), CL40 Fab (green), and E1D1 Fab (orange). The CL59 Fab shown here is based on a generic Fab model as there is no high resolution structure for CL59. The structure was rendered and fitted using UCSF Chimera. (B) Cartoon representation of the 3D reconstruction shown in A, with the heavy chain of each Fab shown in orange and the light chain shown in purple. gHgL is color-coded by individual domains: gL in cyan, gH D-I in blue, gH D-II in wheat, gH D-III in green, and gH D-IV in yellow. The model confirms that the three anti-gHgL Fabs have nonoverlapping epitopes on gHgL. (C) Surface mapping of the CL59 epitope on gH D-III/D-IV. Complete sequence and CDR assignments for CL40 and CL59 are shown in Fig. S4. The structures shown in B and C were rendered using MacPymol.