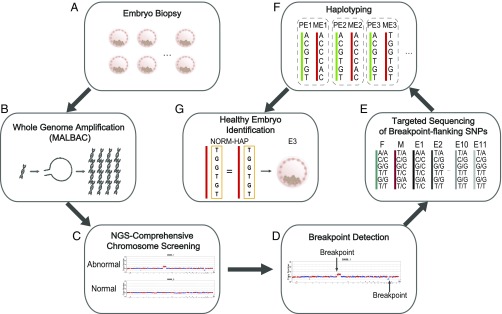
Fig. 1.
Workflow of MaReCs. (A and B) MALBAC-based WGA followed by NGS-based CCS is performed in the biopsy of each embryo. The WGA DNA products generate sufficient DNA that the second round of targeting sequencing can be carried out without a second trophectoderm biopsy of the embryo. Samples are saved for retrospective MaReCs analysis. (C) The embryos with abnormal CNV are identified and excluded from implantation at this stage. (D) Further analysis of the abnormal CNVs of the reference embryo allows identification of the breakpoint of the reciprocal translocation. (E) Multiple SNPs flanking the breakpoint are then targeted and sequenced by NGS for each embryo and the parents. (F) By linkage analysis of the informative SNPs, the haplotypes linked to the normal allele and to the translocation allele of the parental RecT carrier can be mapped separately. (G) Using the haplotype linkage information, the carrier status of the embryos with balanced ploidy can be resolved. Detailed haplotype linkage analysis to resolve the carrier status of embryos is illustrated in Fig. 4. E, embryo; F, father; M, mother; ME, maternal allele of the embryo; PE, paternal allele of the embryo.