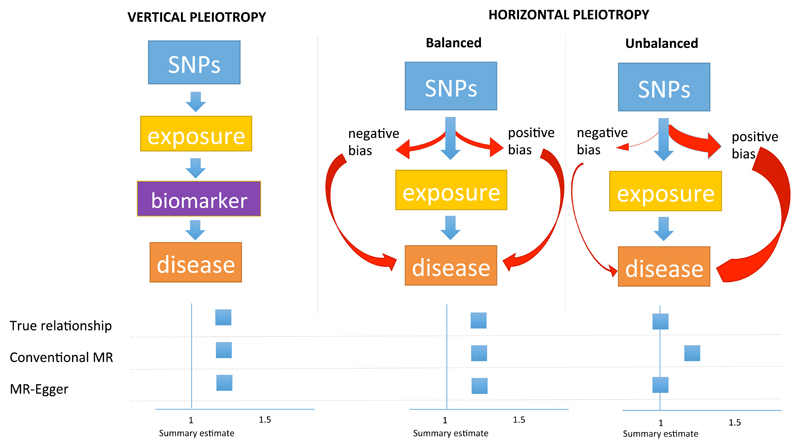
Figure 1. Pleiotropy and the validity of estimates derived from Mendelian randomization.
SNPs are used in a genetic instrument for an exposure to assess the association with risk of disease. For each exposure there is a ‘true relationship’, which we try to approximate from Mendelian randomization. For the purposes of simplicity, conventional MR is compared to MR-Egger.
Vertical pleiotropy explains where the genetic instrument associates with biomarkers (other than the exposure) that are on the causal pathway from exposure through to disease. Horizontal pleiotropy is where the genetic instrument associates with additional traits not on the causal pathway of the exposure of interest. When horizontal pleiotropy is balanced, there should be no bias in the effect derived from MR. In this scenario, the estimate obtained from conventional MR is similar to that from MR-Egger.
When horizontal pleiotropy is unbalanced (also termed ‘directional pleiotropy’), the pleiotropy systematically biases the estimate (which can be exaggerated or diminished) in a naïve analysis using conventional MR. In the example in Figure 1, the unbalanced pleiotropy exaggerates the magnitude of the association. Conventional MR will derive a biased estimate, whereas MR-Egger, correcting for unbalanced pleiotropy, should yield a valid estimate. An example of unbalanced horizontal pleiotropy is the relationship of HDL-C and risk of CAD; the association derived from conventional MR is different to that of MR-Egger with the latter indicating that, once unbalanced pleiotropy is accounted for, there is no effect of HDL-C on risk of CAD (see Figure 3).