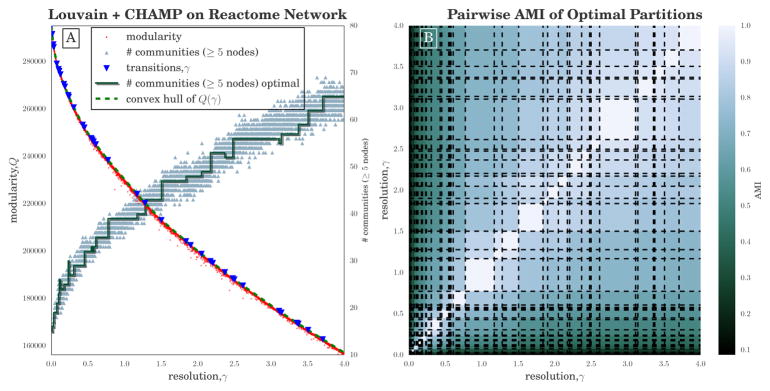
Figure 3.
(A) Modularity Q(γ) given by Equation (1) v. resolution parameter γ for 20, 000 runs (25% of results shown) of Louvain [42,47] on the Human Protein Reactome network [54]. Small, grey triangles indicate the number of communities that include ≥ 5 nodes in each run, while the dark green step function shows the number in the optimal partition in each domain. The dashed green curve is the piecewise-linear modularity function for the optimal partitions, with the transition values marked by blue triangles; (B) Pairwise AMI between all partitions in the admissible subset identified by CHAMP, arranged by their corresponding γ-domains of optimality.