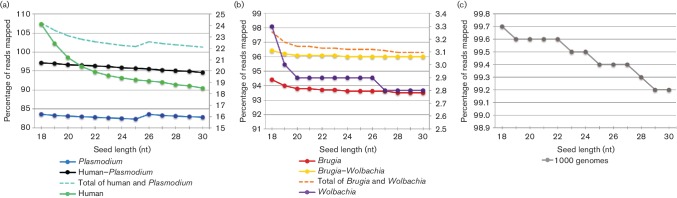
Fig. 1.
The percentage of mapped read pairs for all bwa-mem datasets. The percentage of mapped read pairs for all datasets against all references is shown for each seed length for (a) the human–Plasmodium dataset, (b) the Brugia–Wolbachia dataset and (c) the human-only dataset. The sequencing reads were aligned to the reference genome of each species separately, as well as a combined reference of both genomes, as indicated in the legend. Mappings to the reference genome of the minority member in the sample (human for the Plasmodium–human dataset and Wolbachia for the Brugia–Wolbachia dataset) were plotted on the secondary y-axis on the right, while all others were plotted on the primary y-axis on the left. The sum of the mappings to individual reference genomes is illustrated with a dashed line to enable comparisons.