Abstract
Emerging evidence supports a key role for the gut microbiota in metabolic diseases, including type 2 diabetes (T2D) and obesity. The dipeptidyl peptidase-4 inhibitor vildagliptin is highly efficacious in treating T2D. However, whether vildagliptin can alter the gut microbiome is still unclear. This study aimed to identify whether vildagliptin modifies the gut microbiota structure during T2D treatment. Diabetic Sprague-Dawley (SD) rats were induced by a high-fat diet and streptozotocin injection (HFD/STZ). Diabetic rats were orally administered a low dose of vildagliptin (LV, 0.01 g/kg/d vildagliptin), high dose of vildagliptin (HV, 0.02 g/kg/d vildagliptin), or normal saline for 12 weeks. Fasting blood glucose, blood glucose after glucose loading, and serum insulin levels were significantly reduced in the LV and HV groups compared with those in the T2D group. The serum GLP-1 level increased more in the vildagliptin-treated group than in the T2D group. Pyrosequencing of the V3-V4 regions of 16S rRNA genes revealed that vildagliptin significantly altered the gut microbiota. The operational taxonomic units (OTUs) and community richness (Chao1) index were significantly reduced in the vildagliptin and diabetic groups compared with those in the control group. At the phylum level, a higher relative abundance of Bacteroidetes, lower abundance of Firmicutes, and reduced ratio of Fimicutes/Bacteroidetes were observed in the vildagliptin-treated group. Moreover, vildagliptin treatment increased butyrate-producing bacteria, including Baceroides and Erysipelotrichaeae, in the diabetic rats. Moreover, Lachnospira abundance was significantly negatively correlated with fasting blood glucose levels. In conclusion, vildagliptin treatment could benefit the communities of the gut microbiota.
Introduction
Type 2 diabetes (T2D) is rapidly increasing across the world. Environmental and genetic factors are considered the key underlying reasons for T2D epidemic. Tremendous attention and efforts have been given to decipher the “true story” of T2D. However, understanding how the location of candidate genes and single nucleotide polymorphisms (SNPs) identified in genome-wide association studies (GWAS) and the interaction between genes and the environment affect T2D is still limited. Increasing evidence supports a key role for the gut microbiota in the development of T2D [1, 2]. Previous studies have shown evidence that a high-fat diet changes the ratio of Firmicutes and Bacteroidetes phyla in the gut [3–5]. Mice fed with a high-fat and high-sugar diet showed a shift in the structure of the gut microbiota even within a single day [6]. More inspiring results came from germ- free animals. Several research groups have announced that germ free mice are resistant to diet-induced obesity and insulin resistance [7]. Transferring the fecal microbiota from ob/ob mice to germ-free mice results in increased body weight and insulin resistance [8, 9]. The following pathways are involved: insulin signaling [2], inflammation [10], glucose homeostasis [11], and increased glucagon-like peptide 1 (GLP-1) and peptide YY (PYY) secretion through short-chain fatty acids (SCFA) stimulation [12]. Therefore, Therapy that modulates the gut microbiota may prevent the growing T2D epidemic [13].
Vildagliptin, a dipeptidyl peptidase-4 inhibitor, was approved by the European Agency in 2008 for the treatment of T2D. This inhibitor avoids the enzymatic degradation of glucagon-like peptide 1 (GLP-1). GLP-1 is secreted from the intestinal L-cells. GLP-1 can stimulate insulin secretion from the pancreatic β-cells into the blood in response to glucose intake, reduced glucagon secretion from pancreatic α-cells, delayed gastric emptying and suppressed appetite, and it can protect pancreatic β-cells from apoptosis and promote β-cell proliferation [14].
Recently, an oral anti-diabetic agent, metformin, was proven to have beneficial effects on the gut microbiome in moderate hyperglycemia [15]. In a clinical trial, vildagliptin decreased the HbA1c and fasting blood glucose levels as an add-on therapy in patients with type 2 diabetes inadequately controlled by metformin [16]. Thus, we proposed our hypothesis that vildagliptin-treated diabetic rats also have beneficial modifications to their gut microbiome. However, studies about the effect of the whole gut microbiota in diabetic rats are limited. Therefore, we employed 454 pyrosequencing to explore the change in gut microbiota in vildagliptin-treated diabetic rats. The purpose of this research was to identify the important bacteria in the gut that could contribute to treating diabetes.
Material and methods
Generation of diabetic rats and drug treatment
This study was undertaken with the approval of the Animal Care Committee of the Peking Union Medical Hospital Animal Ethics Committee (Project XHDW-2015-0051, 15 Feb 2015), and all efforts were made to minimize suffering. Five-week-old male Sprague-Dawley (SD) rats (158.3 ± 14.8 g) were acquired from the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China, SCXK-2014-0013). All rats were caged at 24 ± 1 oC with lights on from 6:00 a.m. to 6:00 p.m.
The rats were fed a standard diet (kcal %: 10% fat, 20% protein, and 70% carbohydrate; 3.85 kcal/gm) or a high-fat diet (kcal %: 45% fat, 20% protein, and 35% carbohydrate; 4.73 kcal/gm, Research Diet, New Brunswick, NJ, USA). After exposure to the respective diets for 4 weeks, the rats were injected with STZ (30 mg/kg body weight, i.p.) or saline after being deprived of food for 10 h, and then, they continued with their original diet through the whole experimentation period. Fasting blood glucose (FBG) > 11.1 mmol/L was determined to be the standard definition for the type 2 diabetes model (HFD/STZ).
The diabetic rats were randomly divided into three subgroups: T2D treated with vehicle only (HFD/STZ, n = 6), T2D treated with 0.01 g vildagliptin/kg body weight/day (LV, n = 6), and T2D treated with 0.02 g vildagliptin/kg body weight/day (HFD/STZ+HV, n = 6). The normal diet-fed rats were orally administered vehicle only (NC, n = 6) or 0.02 g vildagliptin (NC+HV). After 12 weeks of treatment, fresh stools were collected by stimulating the anus in the NC, HFD/STZ, and HV group and immediately stored at -80°C for subsequent analysis. After 10 hours of food deprivation, the rats were anesthetized (ketamine 100 mg/kg i.p., Pharmacia and Upjohn Ltd., Crawley, UK), and then, the rats were sacrificed by decapitation. Blood samples were collected from the intraorbital retrobulbar plexus.
Measurements of body weight, fasting blood glucose, and whole blood HbA1c
Body weight was recorded each month. Fasting blood glucose levels were determined with the Bayer Contour TS glucometer and blood glucose test strips (Bayer, Hamburg, Germany). Whole blood hemoglobin A1c (HbA1c) was measured by a high performance liquid chromatography (HPLC) method (Variant II, Bio-Rad Laboratories, Hercules, CA, USA).
Oral glucose tolerance test (OGTT)
After a 12-week treatment, rats were fasted for 10 h before the glucose tolerance test. The oral glucose load was orally administered at 2 g / kg body weight. Tail blood glucose levels were measured at 30, 60 and 120 minutes after the glucose load. The area under the curve (AUC) was obtained by the linear trapezoid method [17].
Measurement of serum insulin, GLP-1, IL-6, and HOMA-IR
Serum insulin, GLP-1 and IL-6 levels were determined by using an ELISA method (Millipore, Bellerica, MA, USA). The assessment of insulin resistance, the homeostasis model assessment of insulin resistance (HOMA-IR), was calculated from the following formula: FBG (mmol/L) x fasting serum insulin (μIU/mL) / 22.5.
Fecal DNA extraction and 454 pyrophosphate sequencing
Total DNA was obtained from frozen feces using QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). The 16S rRNA genes were amplified using the 341F/806R primer set targeting the V3-V4 region (for 341F 5’-CCTAYGGGRBGCASCAG-3’; for 806R, 5’-GGACTACNNGGGTATCTAAT-3’). Purification of the amplified PCR products was conducted by using a QIAquick PCR purification kit (Qiagen, Hilden, Germany). Bacterial 16S rRNA pyrosequencing was performed on an Illumina HiSeq 2500 platform (Norcross, GA, USA).
After merging, the reads were analyzed by quality filtering, including removing sequences with an average quality score < 20 over a 50 bp sliding window, sequences shorter than 200 bp, sequences with homopolymers longer than six nucleotides, and sequences with ambiguous base calls or incorrect primer sequences. High quality reads were then annotated to operational taxonomic units (OTUs, 97% identity) using UPARSE version 7.1 [18], followed by the selection of representative sequences using QIIME software (version 1.7.0, Quantitative Insights into Microbial Ecology) [19]. OTUs were annotated with taxonomic information based on the RDP classifier version 2.2 [20] algorithm by using the GreenGene Database [21]. The relative abundance of each OTU was determined at the phylum, class, order, family, genus and species levels. Alpha (within a community) and beta diversity (between communities) were examined using QIIME software. For alpha diversity, Chao1 (community richness) and the Shannon-Wiener index (community diversity) were calculated. For beta diversity, principal coordinates analysis (PCoA) plots were determined by using both weighted and unweighted UniFrac [22]. In addition, linear discriminant analysis (LDA) of the effect size (LEfSe) was used to calculate OTU abundance and determine the differences among groups [23].
Data analysis
The data are expressed as the means ± SD. When the data were normal and variances were equal, differences among the groups were analyzed using one-way ANOVA followed by Tukey’s post hoc test. Otherwise, a Kruskal-Wallis with Mann-Whitney test was applied. Spearman’s correlations between bacterial abundance and metabolic index were assessed. LEfSe was performed first based on the nonparametric factorial Kruskal-Wallis (KW) sum-rank test followed by the Wilcoxon Signed-Rank test. The threshold logarithmic LDA score was set at 3.0 and the scores were ranked. A P-value ≤ 0.05 was considered statistically significant. All data were analyzed with Prism version 5.0 for Windows (GraphPad Software, San Diego, CA, USA).
Results
Body weight
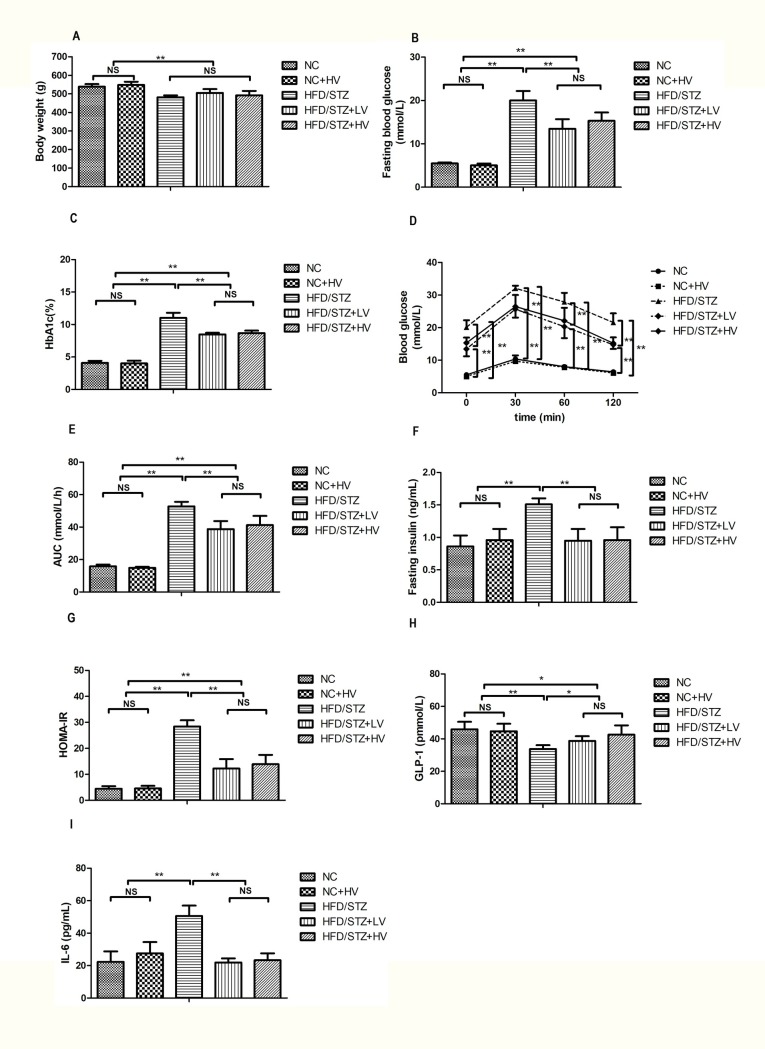
The body weight of diabetic rats was slightly reduced compared with the normal controls (P<0.01, Fig 1A). The body weights of the diabetic rats and vildagliptin-treated rats did not significantly differ (Fig 1A).
Fig 1. Effect of vildagliptin treatment on body weight, blood glucose, HbA1c, serum insulin, HOMA-IR, GLP-1, and IL-6.
(A) body weight, (B) fasting blood glucose, (C) HbA1c, (D) glucose tolerance, (E) area under the curve of blood glucose, (F) serum insulin, (G) HOMA-IR, (H) GLP-1, (I) IL-6. Data was represented as mean ± SD. n = 6 in each group. * P<0.05, ** P<0.01, NS not significant.
Fasting blood glucose, HbA1c and glucose tolerance
Fasting blood glucose and HbA1c levels in the HFD/STZ rats were significantly increased compared with those of the normal controls (P<0.01, Fig 1B). Both doses of vildagliptin treatment reduced the fasting blood glucose and HbA1c levels (P<0.01, Fig 1B and 1C). The blood glucose level after an oral glucose tolerance test was significantly higher in the diabetic group than in the normal controls (P<0.01, Fig 1D). Vildagliptin treatment reduced the blood glucose levels before and after glucose load, and the area under the curve of blood glucose (P<0.01, Fig 1D and 1E).
Fasting insulin and homeostasis model assessment of insulin resistance (HOMA-IR), GLP-1, and IL-6
Serum insulin level, HOMA-IR, and IL-6 level in the diabetic rats were higher than that in the normal controls (P<0.01, Fig 1F, 1G and 1I), However, the serum GLP-1 level was lower in the diabetic rats than in the normal controls (P<0.01, Fig 1H) Vildagliptin reduced the serum insulin and IL-6 levels, alleviated insulin resistance, and increased serum GLP-1 in diabetic rats (P<0.05, Fig 1F, 1G, 1H and 1I).
Global structural changes in the gut microbiota in response to vildagliptin treatment
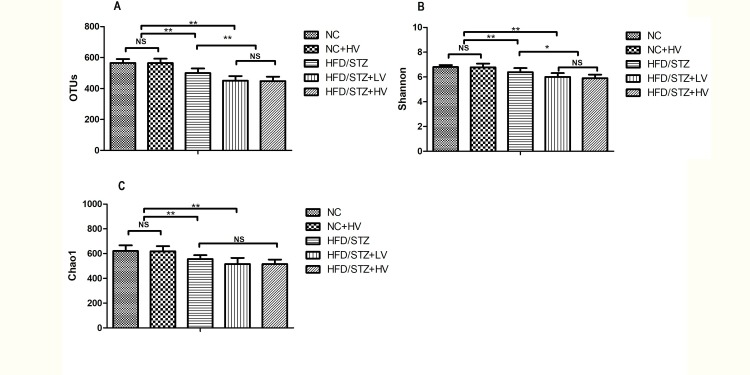
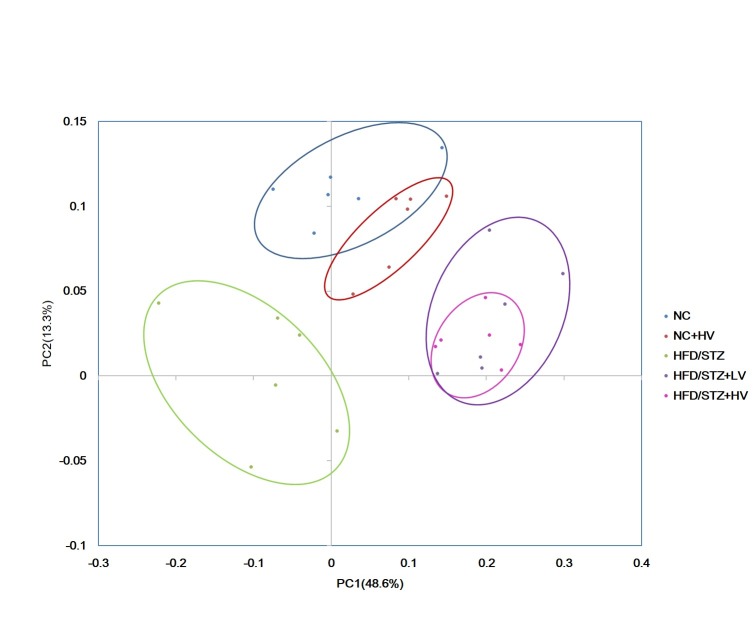
A total of 1,609,238 usable pyrosequencing reads (61,894 unique sequences) were obtained from 30 samples. The 16S sequence data generated in this study were submitted to the NCBI Sequence Read Archive (SRA) database (accession number SRP095711). After quality filtering, 1,289,798 reads (an average of 2213 sequences per sample) were identified as 918 OTUs at the 97% similarity level. Further statistical analysis of the estimators of OTUs and Shannon index showed that the HFD/STZ group had lower microbial diversity than the NC group (P<0.01, Fig 2A and 2B), and vildagliptin treatment reduced the microbial diversity in the HFD/STZ rats (P<0.05, Fig 2A and 2B). The Chao 1 index indicated that the gut microbiota richness in the HFD/STZ group was significantly lower than in the NC group (P<0.01, Fig 2C), however, there was similar richness among HFD/STZ, HFD/STZ+LV and HFD/STZ+HV groups (P>0.05, Fig 2C). Principal coordinates analysis of weighted UniFrac distances performed on the 97% OTU abundance matrix revealed a distinct separation in the beta diversity of the gut microbial communities among the NC, HFD/STZ, and HV groups. However, the NC and NC-HV groups shared some overlap (Fig 3). Alpha and beta diversity analyses of the NC+HV group showed that vildagliptin has little effect on the physiological state.
Fig 2. Pyrosequencing data summary.
(A) number of OTUs, (B) Shannon index, and (C) Chao1 index. Data was represented as mean ± SD. n = 6 in each group. *P<0.05, ** P<0.01, NS not significant.
Fig 3. PCoA plots of weighted UniFrac distances of fecal microbial communities.
The microbial structure of vildagliptin-treated rats differed substantially from that of diabetic rats
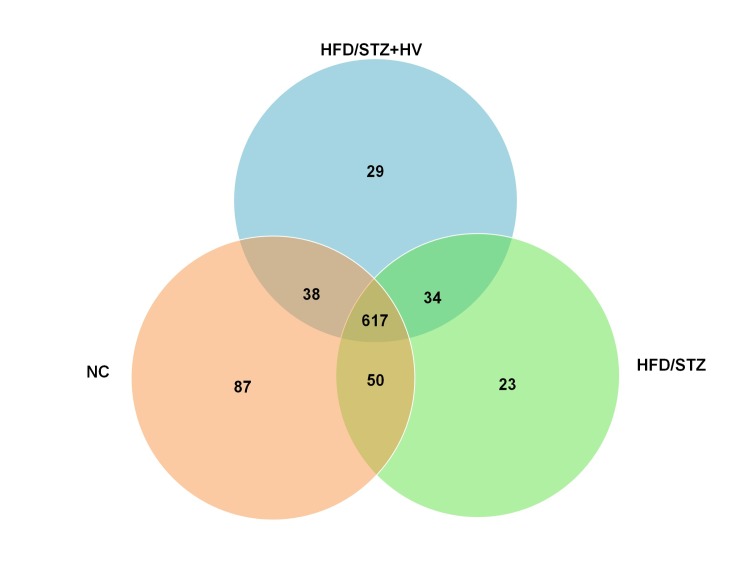
To further explore the similarities and differences among normal, diabetic and vildagliptin-treated rats, we explored the similarities and distinctions in species distribution in the NC, HFD/STZ, and HFD/STZ+HV groups. As shown in Fig 4, we found that there were 617 shared species among the three groups, accounting for approximately three-fourths of the OTUs in each group. It is noteworthy that only 87 species were found in the NC group, 23 OTUs in the HFD/STZ group, and 29 OTUs in the HV group. At the genus level, the unique OTUs in the HV group were Silanimonas (from the Xanthomonadaceae family), Coprobacillus (from the Erysipelotrichaceae family), Parabacteroides (from the Porphyromonadaceae family), Psychrosphaera (from the Pseudoalteromonadaceae family), Peptococcus (from the Peptococcaceae family), Anaerotruncus (from the Ruminococcaceae family), Gardnerella (from the Bifidobacteriaceae family), Ruminococcaceae_UCG-010 (from the Ruminococcaceae family), Acinetobacter (from the Moraxellaceae family), and Alistipes (from the Rikenellaceae family).
Fig 4. Shared OUT analysis of the different groups.
Venn diagram showing the unique and shared OTUs (3% distance level) in the different groups.
The microbiota in the HFD/STZ rats featured a high percentage of Firmicutes and both a low percentage of Bacteroidetes and a low ratio of Bacteroidetes: Firmicutes (P<0.01, S1 Table, Fig 5,). This composition was modified by vildagliptin treatment (P<0.01). Moreover, vildagliptin treatment reduced the relative abundance of Tenericutes and Elusimicrobia (P<0.05, S1 Table, Fig 5).
Fig 5. Heat map of the relative abundance (log10 transformation) of specific taxa in the fecal microbiota of rats in phyla level.
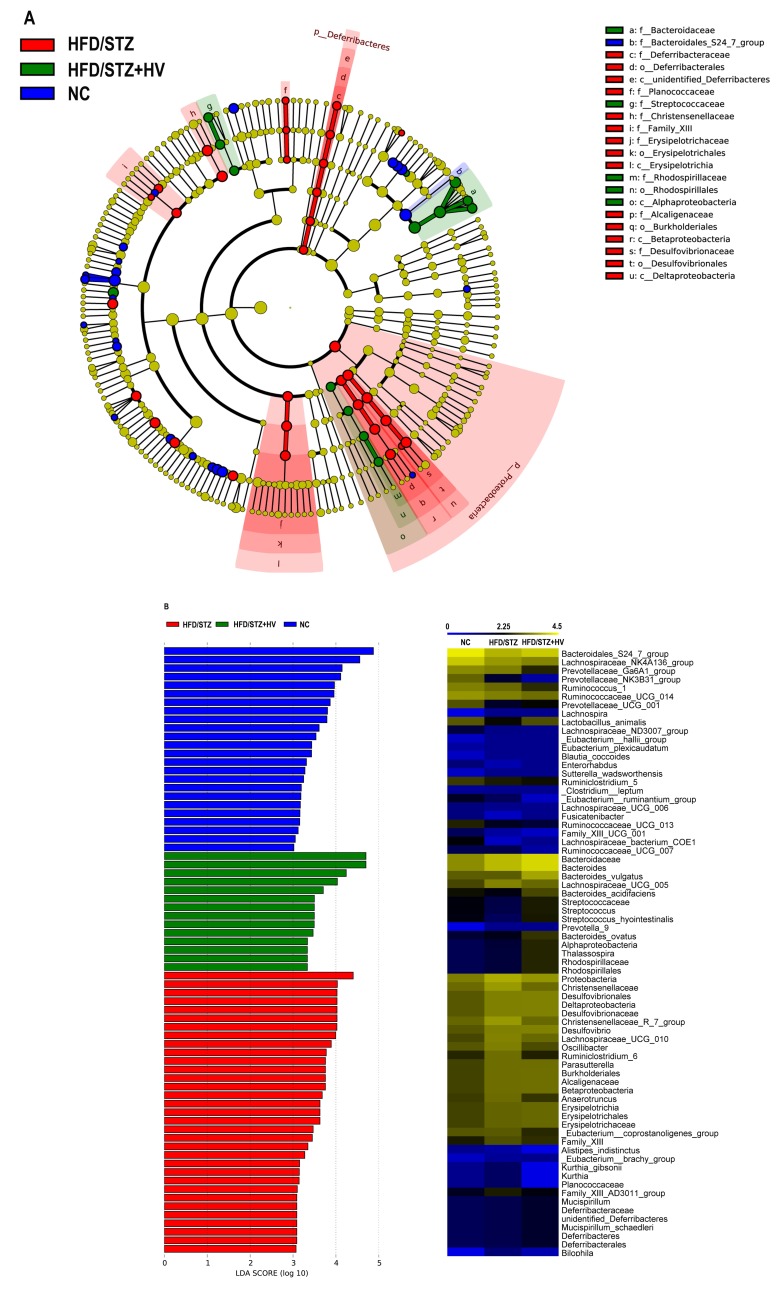
There were significant alterations in the structure of intestinal bacteria species among the NC, HFD/STZ, and HFD/STZ+HV groups. A cladogram representation of the microbiota structure of these three conditions, the predominant bacteria in each condition performed by LEfSe, and the greatest differences in taxa among the three communities are presented in Fig 6 and S2 Table. We further discovered the key variables that separated the gut microbiota under vildagliptin treatment and identified 15 phylotypes as high-dimensional biomarkers. Four of these phylotypes were increased, and 11 phylotypes were decreased under vildagliptin treatment compared with the HFD/STZ group. The enriched phylotypes were the Streptococcaceae family (P<0.01), the genus Streptococcus within the family Streptococcaceae (P<0.01), the species Bacteroides_acidifaciens within the genera Bacteroides and the family Bacteroidaceae (P<0.01), and the species Streptococcus_hyointestinalis within the genus Streptococcus and the family Streptococcaceae (P<0.01). The decreased phylotypes were mainly the family Ruminococcaceae, such as the genera Oscillibacter (P<0.05), Ruminiclostridium_6 (P<0.05), Anaerotruncus (P<0.01), Eubacterium_coprostanoligenes_group (P<0.05), and Ruminococcaceae_UCG_007 (P<0.05); the family Planococcaceae, such as the genus Kurthia (P<0.05), and the species Kurthia_gibsonii (P<0.05), the family Christensenellaceae, such as the genus Christensenellaceae_R_7_group (P<0.05), and the family Prevotellaceae (P<0.05).
Fig 6. The LEfSe analysis of the gut microbiota in vildagliptin-treated rats differed significantly from that in NC and HFD/STZ group.
(A) Differences are represented in the color of the most abundant group. (B) The left histogram shows the lineages with LDA values of 3.0 or higher as determined by LEfSe. The right heap map shows the relative abundance (log10 transformation) of OTUs.
Identification of the key bacteria associated with the glucose metabolism index
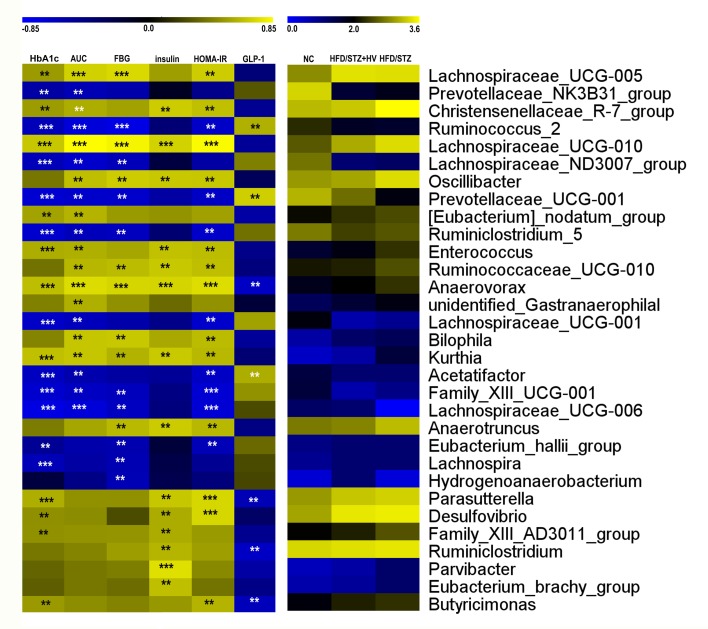
The Spearman correlation of OTU counts (relative abundance) of bacterial genera with glucose metabolism index identified key OTUs that were potentially relevant to blood glucose control in different groups. There were nine genera associated with a decrease in blood glucose levels: Ruminococcus_2, Lachnospiraceae_ND3007_group, Prevotellaceae_UCG-001, Ruminiclostridium_5, Family_XIII_UCG-006, Eubacterium_hallii_group, Lachnospira, and Hydrogenoanaerobacterium (P<0.01). The genera associated with an increase in blood glucose levels were Lachnospiraceae_UCG-005, Lachnospiraceae_UCG-010, Oscillibacter, Ruminococcaceae_UCG-010, Anaerovorax, Kurghia, Bilophila, and Anaerotruncus (P<0.01, S3 Table, S4 Table, Fig 7). In addition, there were fourteen genera positively associated with fasting insulin levels: Parasutterella, Desulfovibrio, Christensenellaceae_R-7_group, Lachnospiraceae_UCG-010, Oscillibacter, Anaerotruncus, Family_XIII_AD3011_group, Ruminococcaceae_UCG-010, Anaerovorax, Kurthia, Parvibacter, and Eubacterium_brachy_group.
Fig 7. Correlation of the gut microbiota with glucose metabolism index.
The left heat map shows the correlation coefficients between the OTUs and various glucose metabolism parameters. The right heat map shows the relative abundance (log10 transformed) of OTUs in the different groups. n = 6 in each group. **P<0.01, *** P<0.001.
Discussion
As expected, our study showed that vildagliptin diminished fasting blood glucose levels, and prevented both glucose intolerance and insulin resistance. In the gut microbiome analysis, we found significant changes among the control, diabetic and vildagliptin-treated groups. PCA revealed distinct fecal microbial communities among the NC, HFD/STZ, and HV groups. The HFD/STZ group had a lower microbial diversity and species richness compared with the NC group. Evidence shows that obesity is associated with reduced microbial diversity and richness [6, 24]. Dietary weight loss interventions have been shown to increase the gene richness of the microbiota [25]. In our study, the vildagliptin-treated rats displayed significantly reduced microbial diversity compared with that of the HFD/STZ rats. Metformin and berberine treatment also decreased the gut microbial diversity in a HFD-induced obesity rodent model [26, 27].
Our data showed that the rat gut microbiome was largely dominated by the phyla Firmicutes and Bacteroidetes. HFD/STZ induced diabetes was related to a significant shift in the gut microbiota, with a reduced percentage of Bacteroidetes and Proteobacteria and an increased proportion of the phyla Firmicutes. In addition, the Bacteroidetes/Firmicutes ratio also declined in the HFD/STZ group. Vildagliptin treatment normalized the Bacteroidetes/Firmicutes ratio, increased the abundance of the phylum Bacteroidetes and decreased the abundance of Firmicutes, Tenericutes (which lack a cell wall), and Elusimicrobia. Previous studies also address the ratio of the phyla Bacteroidetes/Firmicutes as an indicator of microbial imbalance related to a high-fat diet [8, 28, 29].
From the Venn diagram, we identified the unique family Erysipelotrichaceae, in vildagliptin treated rats. At the genus level, vildagliptin increased the level of Bacteroides. Moreover, Lachnospira was negatively correlated with FBG and HOMA-IR. Erysipelotrichaceae, Bacteroides, and Lachnospira are SCFA-producing taxa. Short chain fatty acids (SCFAs) are metabolic products of gut microbiota metabolism. The main products of SCFAs are acetate, propionate, and butyrate. SCFAs can stimulate host energy intake in the intestinal tract and is involved in the development of metabolic diseases [30–32]. Moreover, SCFAs could stimulate the secretion of GLP to affect the intestines [33, 34], and reduce gut permeability to inhibit serum lipopolysaccharide (LPS) and inflammatory cytokine levels [35, 36]. The abundance of the SCFA-producing bacteria, Bacteroides, in diabetic patients was reduced compared with subjects with normal glucose tolerance and with prediabetes [37, 38]. Moreover, physically fit subjects have an increased abundance of Erysipelotrichaceae in the gut microbiota [39]. Some oral antidiabetic agents, namely berberine and metformin, and energy-restricted diets could increase the SCFA-producing bacteria, Bacteroides and Lachnospira, in HF diet-induced obese rats and diabetic patients [40, 41].
Our data showed that Prevotellaceae abundance was lower in the vildagliptin treated rats than in the HFD/STZ rats. Brown and coworkers determined that the amount of Prevotellaceae in the gut microbiome was positively related to post-gestational diabetes mellitus (GDM) subjects. Prevotella are mucin-degrading bacteria that may be related to increased gut permeability [42]. Several studies report a negative relationship between the Bacteroides/Prevotella ratio and metabolic diseases, such as obesity [43] and non-alcoholic steatohepatitis [44].
Additionally, in our study, Ruminococcaceae abundance was lower in the vildagliptin-treated rats than in diabetic rats. In another study, there was a reduction in the abundance of Ruminococcaceae in rats treated with a bitter melon formulation (which could reduce fasting blood glucose) compared to the diabetic rats [45]. In addition, in our study, the abundance of Oscillibacter, one species within the taxa Ruminococcaceae, was reduced in vildagliptin-treated rats. In addition, Oscillibacter abundance was positively correlated with FBG, AUC in OGTT, fasting serum insulin, and HOMA-IR. Oscillibacter is positively correlated with gut permeability, which can affect gut barrier integrity [46]. The abundance of Oscillibacter increased under the HFD condition, and the abundance of these species declined after treatment with an inhibitor of the mTOR complex [47].
Our data also showed that the abundance of Desulfovibrio was positively correlated with fasting serum insulin level and HOMA-IR. As gram-negative bacteria, most members of Desulfovibrio are LPS producers [48, 49] and can damage the gut barrier [50]. A HFD can induce a leaky gut and increase bacterial lysis, allowing the LPS of gram-negative bacteria to enter the enterohepatic circulation [51]. LPS can activate the secretion of pro-inflammatory cytokines, leading to insulin resistance and relative metabolic disorders [35]. A previous study found that the family Desulfovibrionaceae was more common in IGT/obese rats, compared to normal controls [52]. Xie et al. found that Desulfovibrio was markedly increased in STZ-HFD-induced nonalcoholic steatohepatitis and was positively correlated with LPS levels [53].
Correlation analysis showed that Bilophila was positively correlated with FBG, AUC in OGTT and HOMA-IR. Recently, some studies have reported a positive relationship between the abundance of Bilophila wadsworthia, a high-fat diet and inflammation status [54, 55]. Our data also showed that vildagliptin could reduce the serum inflammatory cytokine IL-6 in diabetic rats.
Conclusion
Our findings suggest that vildagliptin enriched SCFA-producing bacteria and reduced gut microbial diversity for improved gastrointestinal health and could eventually mediate their beneficial effects on the host, particularly in diabetes. The changes in the gut microbiota are associated with various metabolic biomarkers. We propose that modifying the specific structure and characteristics of the gut microbiota by vildagliptin treatment might become a therapeutic strategy for metabolic diseases, including T2D.
Supporting information
(PDF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We are very grateful to Beijing Compass Biotechnology Company for excellent technical assistance with 16s sequencing experiments.
Data Availability
Data are available from the SRA database (SRP095711).
Funding Statement
This work was supported by National Key Research and Development Program of China (2016YFA0101002), National Natural Science Foundation of China (81170736, 81570715), National Natural Science Foundation for Young Scholars of China (81300649), China Scholarship Council Foundation (201308110443), PUMC Youth Fund (33320140022), China Diabetes Young Scientific Talent Research Funding, Diabetes Research Fund from Clinical Research of Chinese Medical Association and Chinese Diabetes Society (12030500350), Fundamental Research Funds for the Central Universities, and the Scientific Activities Foundation for Selected Returned Overseas Professional of Human Resources and Social Security Ministry. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Baothman OA, Zamzami MA, Taher I, Abubaker J, Abu-Farha M. The role of Gut Microbiota in the development of obesity and Diabetes. Lipids Health Dis. 2016;15:108 doi: 10.1186/s12944-016-0278-4 ; PubMed Central PMCID: PMCPMC4912704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tai N, Wong FS, Wen L. The role of gut microbiota in the development of type 1, type 2 diabetes mellitus and obesity. Rev Endocr Metab Disord. 2015;16(1):55–65. doi: 10.1007/s11154-015-9309-0 ; PubMed Central PMCID: PMCPMC4348024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esteve E, Ricart W, Fernandez-Real JM. Gut microbiota interactions with obesity, insulin resistance and type 2 diabetes: did gut microbiote co-evolve with insulin resistance? Curr Opin Clin Nutr Metab Care. 2011;14(5):483–90. doi: 10.1097/MCO.0b013e328348c06d . [DOI] [PubMed] [Google Scholar]
- 4.Kubeck R, Bonet-Ripoll C, Hoffmann C, Walker A, Muller VM, Schuppel VL, et al. Dietary fat and gut microbiota interactions determine diet-induced obesity in mice. Mol Metab. 2016;5(12):1162–74. doi: 10.1016/j.molmet.2016.10.001 ; PubMed Central PMCID: PMCPMC5123202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holtcamp W. Gut check: do interactions between environmental chemicals and intestinal microbiota affect obesity and diabetes? Environ Health Perspect. 2012;120(3):A123 doi: 10.1289/ehp.120-a123a ; PubMed Central PMCID: PMCPMC3295372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14 doi: 10.1126/scitranslmed.3000322 ; PubMed Central PMCID: PMCPMC2894525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabot S, Membrez M, Bruneau A, Gerard P, Harach T, Moser M, et al. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010;24(12):4948–59. doi: 10.1096/fj.10-164921 . [DOI] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31. doi: 10.1038/nature05414 . [DOI] [PubMed] [Google Scholar]
- 9.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–5. doi: 10.1073/pnas.0504978102 ; PubMed Central PMCID: PMCPMC1176910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes. 2012;3(4):279–88. doi: 10.4161/gmic.19625 ; PubMed Central PMCID: PMCPMC3463487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829 doi: 10.1038/ncomms2852 ; PubMed Central PMCID: PMCPMC3674247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Curr Pharm Des. 2009;15(13):1546–58. . [DOI] [PubMed] [Google Scholar]
- 13.Samah S, Ramasamy K, Lim SM, Neoh CF. Probiotics for the management of type 2 diabetes mellitus: A systematic review and meta-analysis. Diabetes Res Clin Pract. 2016;118:172–82. doi: 10.1016/j.diabres.2016.06.014 . [DOI] [PubMed] [Google Scholar]
- 14.Ross SA, Ekoe JM. Incretin agents in type 2 diabetes. Can Fam Physician. 2010;56(7):639–48. ; PubMed Central PMCID: PMCPMC2922799. [PMC free article] [PubMed] [Google Scholar]
- 15.Maniar K, Moideen A, Mittal A, Patil A, Chakrabarti A, Banerjee D. A story of metformin-butyrate synergism to control various pathological conditions as a consequence of gut microbiome modification: Genesis of a wonder drug? Pharmacol Res. 2017;117:103–28. doi: 10.1016/j.phrs.2016.12.003 . [DOI] [PubMed] [Google Scholar]
- 16.Bosi E, Camisasca RP, Collober C, Rochotte E, Garber AJ. Effects of vildagliptin on glucose control over 24 weeks in patients with type 2 diabetes inadequately controlled with metformin. Diabetes Care. 2007;30(4):890–5. doi: 10.2337/dc06-1732 . [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, Sun X, Xiao X, Zheng J, Li M, Yu M, et al. Maternal Chromium Restriction Leads to Glucose Metabolism Imbalance in Mice Offspring through Insulin Signaling and Wnt Signaling Pathways. Int J Mol Sci. 2016;17(10). doi: 10.3390/ijms17101767 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–8. doi: 10.1038/nmeth.2604 . [DOI] [PubMed] [Google Scholar]
- 19.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303 ; PubMed Central PMCID: PMCPMC3156573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–7. doi: 10.1128/AEM.00062-07 ; PubMed Central PMCID: PMCPMC1950982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–72. doi: 10.1128/AEM.03006-05 ; PubMed Central PMCID: PMCPMC1489311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005 ; PubMed Central PMCID: PMCPMC1317376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60 doi: 10.1186/gb-2011-12-6-r60 ; PubMed Central PMCID: PMCPMC3218848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–6. doi: 10.1038/nature12506 . [DOI] [PubMed] [Google Scholar]
- 25.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500(7464):585–8. doi: 10.1038/nature12480 . [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One. 2012;7(8):e42529 doi: 10.1371/journal.pone.0042529 ; PubMed Central PMCID: PMCPMC3411811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H, Ko G. Effect of metformin on metabolic improvement and gut microbiota. Appl Environ Microbiol. 2014;80(19):5935–43. doi: 10.1128/AEM.01357-14 ; PubMed Central PMCID: PMCPMC4178684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137(5):1716–24 e1-2. doi: 10.1053/j.gastro.2009.08.042 ; PubMed Central PMCID: PMCPMC2770164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy EF, Cotter PD, Healy S, Marques TM, O'Sullivan O, Fouhy F, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59(12):1635–42. doi: 10.1136/gut.2010.215665 . [DOI] [PubMed] [Google Scholar]
- 30.Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. 2008;105(43):16767–72. doi: 10.1073/pnas.0808567105 ; PubMed Central PMCID: PMCPMC2569967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alles MS, Hautvast JG, Nagengast FM, Hartemink R, Van Laere KM, Jansen JB. Fate of fructo-oligosaccharides in the human intestine. Br J Nutr. 1996;76(2):211–21. . [DOI] [PubMed] [Google Scholar]
- 32.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40(3):235–43. . [DOI] [PubMed] [Google Scholar]
- 33.Tappenden KA, McBurney MI. Systemic short-chain fatty acids rapidly alter gastrointestinal structure, function, and expression of early response genes. Dig Dis Sci. 1998;43(7):1526–36. . [DOI] [PubMed] [Google Scholar]
- 34.Tappenden KA, Drozdowski LA, Thomson AB, McBurney MI. Short-chain fatty acid-supplemented total parenteral nutrition alters intestinal structure, glucose transporter 2 (GLUT2) mRNA and protein, and proglucagon mRNA abundance in normal rats. Am J Clin Nutr. 1998;68(1):118–25. . [DOI] [PubMed] [Google Scholar]
- 35.Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57(6):1470–81. doi: 10.2337/db07-1403 . [DOI] [PubMed] [Google Scholar]
- 36.Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58(8):1091–103. doi: 10.1136/gut.2008.165886 ; PubMed Central PMCID: PMCPMC2702831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, et al. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One. 2013;8(8):e71108 doi: 10.1371/journal.pone.0071108 ; PubMed Central PMCID: PMCPMC3754967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X, Ma C, Han L, Nawaz M, Gao F, Zhang X, et al. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr Microbiol. 2010;61(1):69–78. doi: 10.1007/s00284-010-9582-9 . [DOI] [PubMed] [Google Scholar]
- 39.Estaki M, Pither J, Baumeister P, Little JP, Gill SK, Ghosh S, et al. Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome. 2016;4(1):42 doi: 10.1186/s40168-016-0189-7 ; PubMed Central PMCID: PMCPMC4976518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Zhao Y, Xu J, Xue Z, Zhang M, Pang X, et al. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci Rep. 2015;5:14405 doi: 10.1038/srep14405 ; PubMed Central PMCID: PMCPMC4585776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Candela M, Biagi E, Soverini M, Consolandi C, Quercia S, Severgnini M, et al. Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet. Br J Nutr. 2016;116(1):80–93. doi: 10.1017/S0007114516001045 ; PubMed Central PMCID: PMCPMC4894062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6(10):e25792 doi: 10.1371/journal.pone.0025792 ; PubMed Central PMCID: PMCPMC3197175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59(12):3049–57. doi: 10.2337/db10-0253 ; PubMed Central PMCID: PMCPMC2992765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58(1):120–7. doi: 10.1002/hep.26319 . [DOI] [PubMed] [Google Scholar]
- 45.Zhu Y, Bai J, Zhang Y, Xiao X, Dong Y. Effects of bitter melon (Momordica charantia L.) on the gut microbiota in high fat diet and low dose streptozocin-induced rats. Int J Food Sci Nutr. 2016;67(6):686–95. doi: 10.1080/09637486.2016.1197185 . [DOI] [PubMed] [Google Scholar]
- 46.Lam YY, Ha CW, Campbell CR, Mitchell AJ, Dinudom A, Oscarsson J, et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS One. 2012;7(3):e34233 doi: 10.1371/journal.pone.0034233 ; PubMed Central PMCID: PMCPMC3311621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung MJ, Lee J, Shin NR, Kim MS, Hyun DW, Yun JH, et al. Chronic Repression of mTOR Complex 2 Induces Changes in the Gut Microbiota of Diet-induced Obese Mice. Sci Rep. 2016;6:30887 doi: 10.1038/srep30887 ; PubMed Central PMCID: PMCPMC4965768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loubinoux J, Mory F, Pereira IA, Le Faou AE. Bacteremia caused by a strain of Desulfovibrio related to the provisionally named Desulfovibrio fairfieldensis. J Clin Microbiol. 2000;38(2):931–4. ; PubMed Central PMCID: PMCPMC86253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weglarz L, Dzierzewicz Z, Skop B, Orchel A, Parfiniewicz B, Wisniowska B, et al. Desulfovibrio desulfuricans lipopolysaccharides induce endothelial cell IL-6 and IL-8 secretion and E-selectin and VCAM-1 expression. Cell Mol Biol Lett. 2003;8(4):991–1003. . [PubMed] [Google Scholar]
- 50.Beerens H, Romond C. Sulfate-reducing anaerobic bacteria in human feces. Am J Clin Nutr. 1977;30(11):1770–6. . [DOI] [PubMed] [Google Scholar]
- 51.Chang CJ, Lin CS, Lu CC, Martel J, Ko YF, Ojcius DM, et al. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat Commun. 2015;6:7489 doi: 10.1038/ncomms8489 ; PubMed Central PMCID: PMCPMC4557287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4(2):232–41. doi: 10.1038/ismej.2009.112 . [DOI] [PubMed] [Google Scholar]
- 53.Xie G, Wang X, Liu P, Wei R, Chen W, Rajani C, et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget. 2016;7(15):19355–66. doi: 10.18632/oncotarget.8466 ; PubMed Central PMCID: PMCPMC4991388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. doi: 10.1038/nature12820 ; PubMed Central PMCID: PMCPMC3957428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487(7405):104–8. doi: 10.1038/nature11225 ; PubMed Central PMCID: PMCPMC3393783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Data are available from the SRA database (SRP095711).