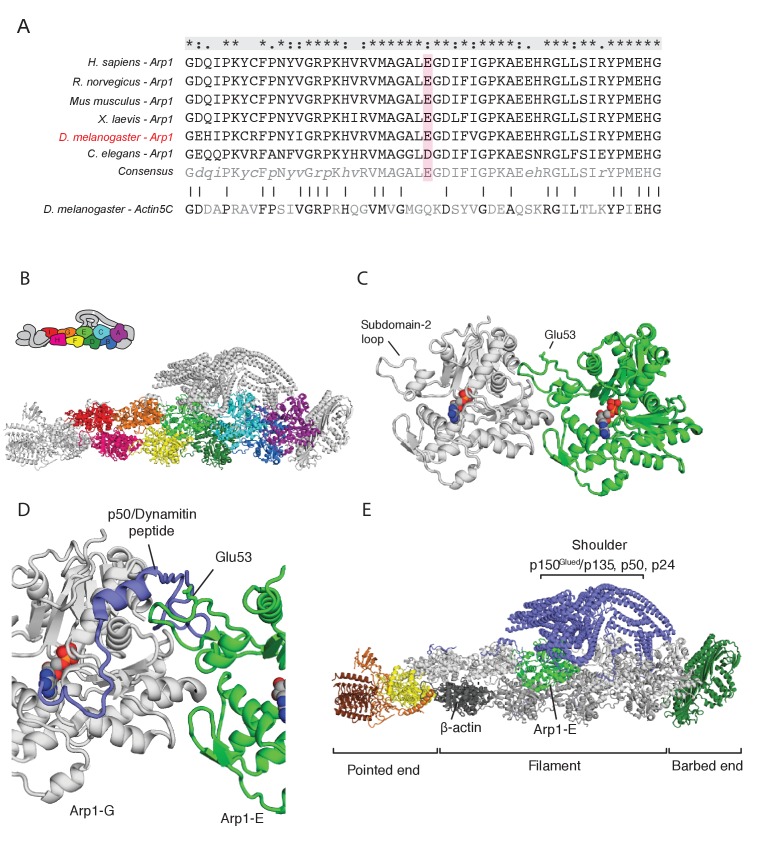
Figure 4. The arp14D2 mutation is predicted to disrupt the interaction between the Arp1 rod and the p150Glued/dynamitin/p24 shoulder domain in the Dynactin complex.
(A) A ClustalX alignment of part of subdomain 2 (amino acids 26–78) in the Arp1 orthologues from different animal species and Drosophila Actin 5C. The orthologues shown are Homo sapiens (human), Rattus norwegicus (rat), Canis familiaris (dog), Xenopus leavis (African clawed toad), Drosophila melanogaster and Caenorhabditis elegans. The pink shading marks the conserved glutamate 53, which is mutated to lysine in the arp14D2 mutant. (B) The structure of dynactin, as determined by Cryo-EM. Eight Arp1 subunits (A, B, C, D, E, F, G, and I) and one actin molecule (H) assemble into two protofilaments that form the rod along the backbone of dynactin. The positions of each subunit are labelled in the drawing at the top and are shown in the same colour in the structure below. (C) A high resolution view of the interface between two Arp1 subunits, showing that E53 does not contribute to the interaction interface, but protrudes laterally. (D) A high resolution view of the interface between Arp1-E subunit and one of the extended peptide domains of p50 dynamitin that anchors the shoulder domain to the Arp1 rod. Glutamate 53 lies at this interface and its substitution by lysine is predicted to disrupt this interaction. (E) A view of the entire dynactin structure showing the positions of Arp1-E (light green) and the p150Glued shoulder domain (blue).