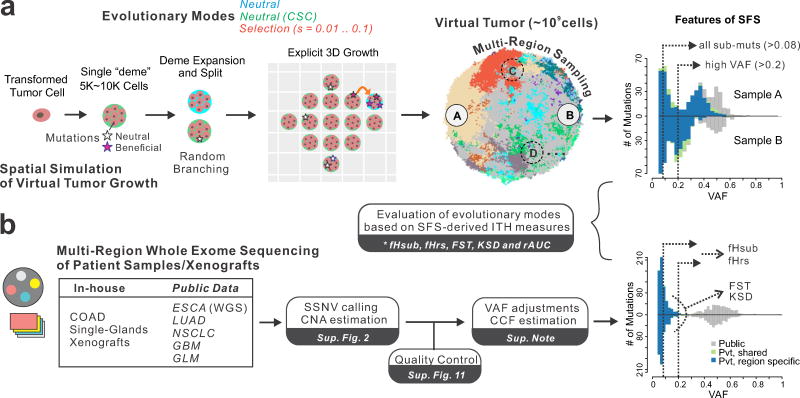
Figure 1. Overview of simulation framework and genomic data analysis pipeline.
(a) Schematic overview of our agent-based computational framework to simulate 3D tumor growth (after transformation) under various modes of evolution, including neutral evolution (null model) and different levels of positive selection, followed by spatial sampling and multi-region sequencing of the virtual tumor. Tumor growth is simulated via the expansion of deme subpopulations within a defined 3D cubic lattice according to explicit rules dictated by spatial constraints, where cells within each deme are well-mixed and grow via a stochastic branching (birth-death) process (Methods and Supplementary Figure 1). By simulating the acquisition of random mutations (neutral or beneficial), tracing the genealogy of each cell as the tumor expands and subsequently virtually sampling and sequencing the ‘final’ virtual tumor as is done experimentally after resection or biopsy, it is possible to evaluate differences in the site frequency spectrum (SFS) under different modes of selection and sampling strategies. Five intra-tumor heterogeneity (ITH) metrics derived from the SFS were employed to distinguish between different evolutionary modes. Sub muts, subclonal mutations. (b) A unified sequencing analysis pipeline based on SSNV calling, copy number estimation, as well as stringent quality control was employed to obtain variant allele frequency (VAF) estimates adjusted for purity and local copy number for seven multi-region sequencing (MRS) datasets derived from patient samples across diverse tissue types. The ITH metrics were similarly computed in patient tumor samples and compared to those observed in virtual tumors under different evolutionary modes.