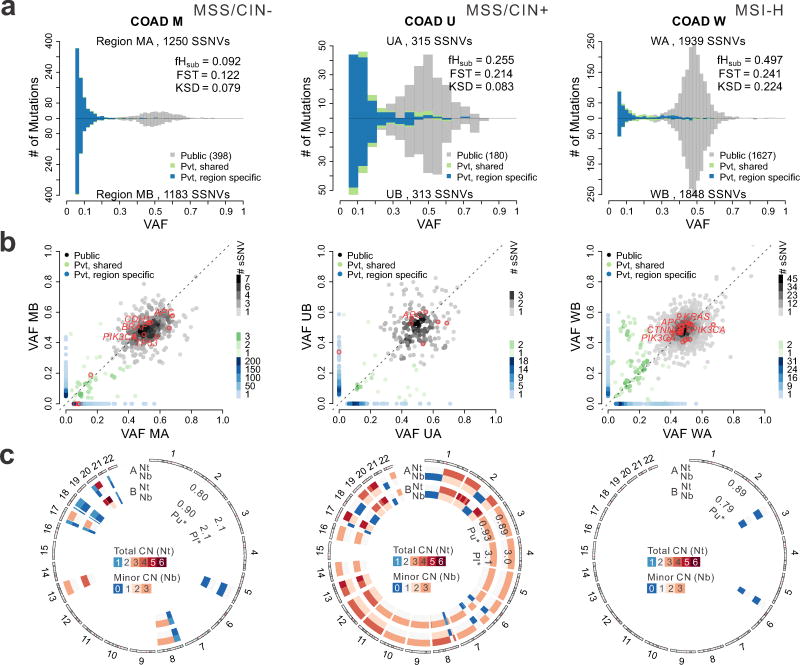
Figure 3. Colorectal tumors exhibit patterns of between-region genetic divergence consistent with effectively-neutral growth or selection.
(a) Pairwise comparison of SFS histograms from each of three bi-sampled colon adenocarcinomas (COADs) representing the major molecular subgroups, including MSI-H (carcinoma W, right), MSS/CIN+ (carcinoma U, middle) and MSS/CIN- (carcinoma M, left). The pairwise histograms illustrate the number of SSNVs detected at a given VAF for the two tumor regions shown above and below the x-axis. SSNVs were classified as Public (gray), Private (Pvt)-shared (green), or Private-region specific (blue). The total number of SSNVs detected in each region and the fHsub, FST, and KSD values are indicated. (b) Scatterplots comparing SSNVs detected in each tumor region at a given VAF. The color of individual SSNV points correspond to that in Panel A and hues reflect the number of SSNVs in a square (0.02 on a side) centered on each SSNV, as depicted in the legend. Nonsilent SSNVs in predicted COAD driver genes are denoted by red circles with known drivers labeled. (c) Circos plot illustrating the predicted absolute total CN (Nt) and minor allele CN (Nb) for each tumor sample. Diploid segments are indicated in white for Nt (two copies) and Nb (one copy), while segments with copy number gain and loss are shown in red and blue, respectively, according to the scale bar. Tumor cell purity (Pu) as well as ploidy (Pl) estimates for each region are indicated on the corresponding concentric rings.