Figure 1.
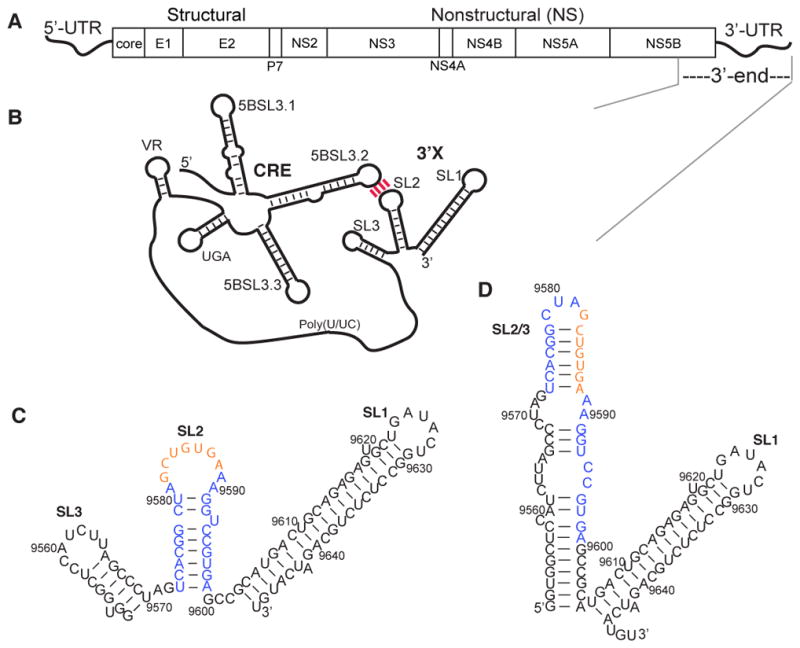
HCV genome and predicted secondary structures for the 3′X region and the 3′-end of the HCV gRNA. (A) The HCV genome contains 5′- and 3′-UTRs and a polyprotein-coding ORF. (B) The 3′X region is proposed to form a long-range interaction with the upstream CRE element via the complementary loop residues between 5BSL3.2 and SL2. (C) The SL2 kissing-loop residues (orange) are readily exposed in the proposed 3SL conformation of the 3′X region. (D) The extended SL2/3 stem–loop structure is shown in the 2SL conformation, which sequesters the SL2 kissing-loop residues.
