Fig. 1. Polη and Polζ can bypass the AP site in vitro.
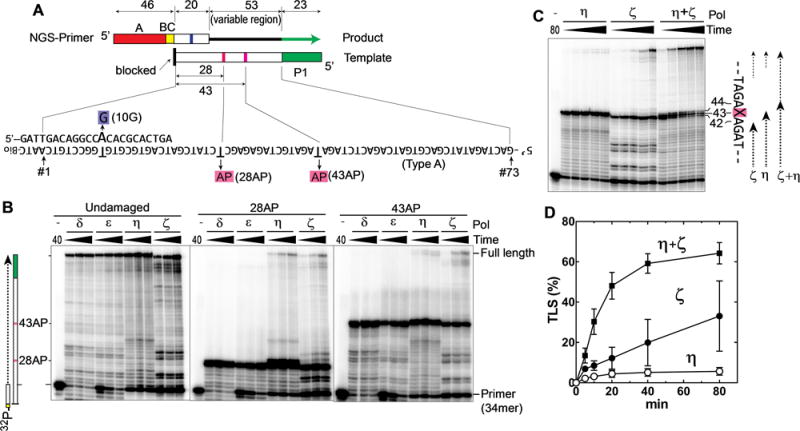
(A) Schematic drawing of Synthetic DNA substrates for TLS reactions. The NGS-primer contained an A-adaptor (red box) and a 6-nt barcode (yellow box, BC) for Ion Torrent PGM. Templates contained a P1-adaptor at the 5′-end (green box) and the complementary sequence to the primer (20-nt-long region) at the 3′ end. The product, which has both adaptors, is recognized by the NGS system that reads the sequences of the 73-nt region (position #1–73), in which the first 20-nt region (position #1–20) are part of the primer, and the remaining 53-nt region (position #21–73; variable region) is extended by yeast DNA polymerases. In addition to the template without lesion (type-A), we used templates with the same sequences but containing an AP site at the position #28 (28AP) or position #43 (43AP). All templates were modified with biotin at the 3′-OH group to prevent extension. The NGS primer had a one-base mismatch at position #10 (10G) to distinguish the primer-extension products from any template-extension product. (B) Primer extension reactions were carried out using the indicated polymerase and 32P-labeled primer (34mer) hybridized to the template with no damage, 28AP, or 43AP. After incubating for 2.5, 5, or 10 min (Polδ and ε), or 10, 20, or 40 min (Polη and ζ), a small portion was withdrawn and analyzed by denaturing polyacrylamide gel. (C) The same TLS reactions as in panel B were carried out on the template containing 43AP by Polη, Polζ, and a mixture of Polη and Polζ, and the reaction was stopped at 5, 10, 20, 40, and 80 min. (D) The TLS efficiency (% of the fully extended products in all DNA molecules that reached the AP site) was calculated from C and repeated experiments (error bar is SD (n=3)).
