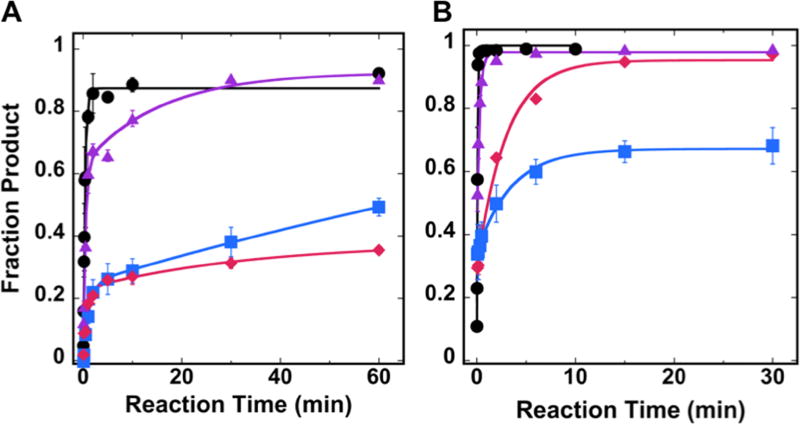
Fig. 3.
Single-turnover kinetic results for hOGG1 and UDG in NCP substrates. Kinetic time courses were performed to evaluate the activity of (A) hOGG1 or (B) UDG on either free duplex DNA (●) or NCP substrates with off-dyad lesions. The NCP substrates contained lesions of varying rotational position: OUT (
 ), MID (
), MID (
 ), or IN (
), or IN (
 ). Experiments were conducted using 20 nM NCP substrate and 0.64 µM glycosylase in 20 mM Tris-HCl (pH 7.6), 25 mM NaCl, 75 mM KCl, 1 mM EDTA, 1 mM DTT, 200 µg/ml BSA. Data for free duplex DNA substrates were fit to a single exponential equation; all NCP data were fit to a double exponential equation. Error bars represent the standard error (n=4).
). Experiments were conducted using 20 nM NCP substrate and 0.64 µM glycosylase in 20 mM Tris-HCl (pH 7.6), 25 mM NaCl, 75 mM KCl, 1 mM EDTA, 1 mM DTT, 200 µg/ml BSA. Data for free duplex DNA substrates were fit to a single exponential equation; all NCP data were fit to a double exponential equation. Error bars represent the standard error (n=4).