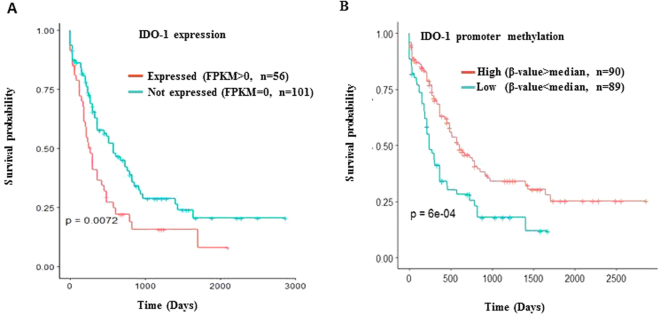
Figure 2.
TCGA database of AML patients downloaded from the UCSC website. (A) RNA seq and Illumina 450 K methylation array data was obtained and the FKPM value was used to divide the external cohort into two groups for survival analysis. Higher FKPM >0 correlates with a poor OS (*p = 0.0072). (B) For methylation array data, one of the three CpG sites assigned to IDO-1 (*cg10262052) was used to divide the cohort into two groups based on the median β-value. Higher median β-values of IDO-1 promoter methylation correlated with better outcomes (*p = 6e-04).