Abstract
Background
Intracellular calcium is an important ion involved in the regulation and modulation of many neuronal functions. From regulating cell cycle and proliferation to initiating signaling cascades and regulating presynaptic neurotransmitter release, the concentration and timing of calcium activity governs the function and fate of neurons. Changes in calcium transients can be used in high-throughput screening applications as a basic measure of neuronal maturity, especially in developing or immature neuronal cultures derived from stem cells.
Results
Using human induced pluripotent stem cell derived neurons and dissociated mouse cortical neurons combined with the calcium indicator Fluo-4, we demonstrate that PeakCaller reduces type I and type II error in automated peak calling when compared to the oft-used PeakFinder algorithm under both basal and pharmacologically induced conditions.
Conclusion
Here we describe PeakCaller, a novel MATLAB script and graphical user interface for the quantification of intracellular calcium transients in neuronal cultures. PeakCaller allows the user to set peak parameters and smoothing algorithms to best fit their data set. This new analysis script will allow for automation of calcium measurements and is a powerful software tool for researchers interested in high-throughput measurements of intracellular calcium.
Keywords: Calcium imaging software, High-content screening, Human stem cells, Calcium imaging
Background
Intracellular calcium ([Ca2+]) regulates key neuronal cell cycle and signaling pathways, including development and proliferation, axon guidance, presynaptic neurotransmitter release, and postsynaptic activity-dependent synaptic plasticity [1–4]. Dysregulation of [Ca2+] concentration has been associated with multiple neurodevelopmental, neuropsychiatric, and neurodegenerative diseases, including Alzheimer disease, autism spectrum disorder, and schizophrenia [5–9]. For extensive reviews of the function of Ca2+ in neuronal cells please see [10, 11]. The combination of Ca2+ imaging with the use of human induced pluripotent stem cells (hiPSCs) provides a very powerful toolset for interrogating the role of subsets of neurons and neural circuits in patient-specific disease models. Besides describing the basal function of hiPSC-derived neurons, Ca2+-based imaging can test the hypothesis that hiPSC-based models accurately recapitulate the genetic background and early developmental physiology of the individuals from whom they are harvested [8, 12–14].
The advent of high-throughput imaging technology, such as ThermoFisher’s Array Scan platform (ThermoFisher Scientific Waltham, MA, USA), combined with the use of hiPSC-based methods can generate large amounts of raw Ca2+ imaging data. Critical to accurate measurements of cellular Ca2+ is the analytics platform through which the raw data is processed. There are currently a number of high-powered Ca2+ analysis programs that have a wide breadth of functionality addressing this problem (Table 1). For example, FluroSNNAP is a program that allows for semi-automated analysis of Ca2+ transient event detection [15]. FluroSNNAP also has the ability to use local and global synchronization analysis to identify related neurons in a network and graph functional connectivity. In order to assure unbiased analysis of cellular Ca2+ spikes, [16] developed a MATLAB based program that focuses on single and multiple transient responses by giving quantifiable parameters for time of onset after stimulus application, amplitude of response, response duration, area under the curve, and decay time constant among other measurements. For looking at large number of cells, NeuroCa is able to process ~ 1000 Ca2+ spikes from a given recording field. This software can allow for the mapping of functional connectivity of large networks of cells by mapping the spatiotemporal Ca2+ spike patterns amongst the network [17]. Another MATLAB-based program developed by [18] is designed to distinguish individual cells in densely populated cultures and measure their Ca2+ activity. The program does this using a segmentation algorithm combined with time-series analysis to measure Ca2+ transients. Finally, SIMA is a Python-based open source software package designed for 2-photon Ca2+ imaging experiments. SIMA is able to efficiently correct for motion artifacts as well as background noise while applying segmentation and extraction of Ca2+ transients from in vitro and in vivo tissue [19, 20].
Table 1.
Comparison of software packages that measure intracellular calcium activity
| Software package | Major features of calcium dynamics | References |
|---|---|---|
| SIMA | Amplitude Time of onset Area under the curve Frequency Activation and decay time |
[19, 20] |
| FluoroSNNAP | Amplitude Functional connectivity Synchronization analysis Correlation coefficient Frequency Activation and decay time Wavelet-based detection |
[15] |
| NeuroCa | Amplitude Frequency Background correction Analysis of ~ 1000 cells Network dynamics |
[17] |
| PeakCaller | Frequency Synchronization index Correlation coefficient Event rise and fall time |
|
| MATLAB dynamics | Amplitude Frequency Time of onset after stimulus Area under the curve Response duration Full width half max Decay time constant |
[16] |
| MATLAB script | Amplitude Frequency Automated detection of single cells in dense cultures Background correction |
[18] |
There are a number of software packages designed to measure Ca2+ transients in cells, and each program has strengths and weaknesses. Nonetheless, many research labs still use the PeakFinder MATLAB script to do simple peak detection of fluorescent signals from cell populations. In light of having many options for the researcher to choose from when analyzing Ca2+ activity in neurons, we have developed PeakCaller-an enhanced version of PeakFinder.
PeakFinder is a MATLAB script which automates the identification of “peaks” of independent variables over time, and is commonly used in the quantification of Ca2+ signaling. However, PeakFinder has several limitations that can result in an increase in type I and type II errors. To address these limitations, our group has developed PeakCaller, a MATLAB GUI interface and algorithm. In the following manuscript, we present empirical evidence of the efficiency of PeakCaller by demonstrating how the use of PeakCaller reduces the occurrence of type I and type II errors. We test this hypothesis across multiple Ca2+ indicators and analysis platforms. Finally, we provide a step-by-step guide to using PeakCaller, and a detailed justification of the algorithms used in PeakCaller.
Implementation
hiPSC-derived cortical neuron cultures
HIP™ Neural Stem Cells (MTI-GlobalStem, Gaithersburg, MD) were cultured according to the manufacturer’s specification. Cells were cultured in GlobalStem Expansion Media (Neurobasal medium)(Life Technologies) supplemented with 2 mM Glutamax (Gibco), 2% B-27 supplement (Gibco), 1X non-essential amino acids (Gibco), and 20 ng/mL FGF-2 on 1:200 Geltrex-coated 6 well culture dishes and incubated at 37 °C/5% CO2. After a 7–10 day recovery period, HIP™ Neural Stem Cells were disassociated from the culture dishes using pre-warmed Accutase (Gibco) and plated at 120,000 cell/well onto Perkin-Elmer 24 well glass-bottom culture plates. Plates were coated with 1X polyornithine (Gibco) for 1 h at 37 °C/5% CO2 and 1:333 Laminin (Life Technologies) for 24 h at 37 °C/5% CO2. The cells were directed toward a neuronal fate using Global Stem maintenance media (NeuralQ Basal Medium, GlobalStem), 2% GS21 Supplement (GlobalStem), and 0.5 mM l-Alanine/l-Glutamine. Three-quarters media change was performed every other day. Cells were kept in these culture conditions until day 30 post differentiation.
Dissociated mouse cortical neuron culture
Cortices from postnatal day 0–0.5 (P0-0.5) mice (C57BL/6J) were dissected out according to procedure described in [21]. Tissue was dissociated for 70–90 min at 37 °C using 10 U/mL Papain (Worthington, Lakewood, NJ) in HBSS containing 0.2 mg/mL l-cysteine (Sigma), 1 mM Pyruvate, 10 mM HEPES, 100 U/mL Penicillin, 100 mg/mL Streptomycin. Thereafter, dissociated tissue was rinsed twice in HBSS solution, with an addition of 10 U/mL DNase (Sigma) in the first wash, and twice in NB-FBS (Neurobasal A medium supplemented with 2% B-27 supplement, 2 mM l-glutamine, 100 U/mL Penicillin, 100 mg/mL Streptomycin and 10% FBS) (Sigma). To obtain a single cell suspension, tissue was triturated in NB-FBS using a P1000 pipette. For Ca2+ imaging experiments, cells were plated onto glass bottom Fluorodishes (World Precision Instruments, Sarasota, FL) coated with 20 mg/mL poly-d-lysine in 20 mM HEPES solution, at a concentration of 1.5 × 106 cells/mL in NB-FBS. Plating medium was exchanged to serum-free NB culture medium after 3–15 h. Cells were maintained in culture for 14 days in vitro (DIV) at 37 °C/5% CO2. All cell culture reagents were from Gibco/Invitrogen (ThermoFisher) unless otherwise stated.
Fluo-4 Ca2+ Assay and pharmacological interrogation
Both hiPSC-derived cortical neurons and dissociated mouse cortical neurons were subjected to the Fluo-4 Direct Ca2+ Assay (ThermoFisher). Assay reagents were prepared according to manufacturer’s protocol and Fluo-4 was added 1:1 with cell culture media. Cells were incubated at 37 °C/5% CO2 for 30 min prior to imaging. Cells were imaged using the Zeiss Spinning disk confocal microscope. Cells were recorded for 10 min at 1 frame every 600 ms (1000 frames total) at 488 nm. Zen Blue software was used to manually designate ‘Regions of Interest’ (ROI) around cell bodies that showed active Ca2+ flux. Raw mean fluorescent data was exported via *.txt file for each ROI. To test how the PeakCaller script would perform under pharmacological conditions resulting in increased neuronal excitation as compared to PeakFinder, we exposed hiPSC-derived cortical neuronal cultures to Fluo-4 for 30 min then added 50 μM of the GABAA channel inhibitor picrotoxin (PTX) (Tocris) to the culture media. Cells were imaged for 40 min (4 × 10 min recordings) and ROIs were selected as described above. The same ROIs (cell bodies) were traced for the duration of the assay. Where indicated, some human iPSC-derived neurons were incubated with Adeno-associated Virus containing vectors for the Ca2+ indicator GCamp6 (AAV-GCamp6, University of North Carolina Vector Core). After incubation, neurons were subjected to imaging using a Zeiss LSM Duo at a rate of 10 Hz (University Of Maryland, School Of Medicine Confocal Microscopy Core).
In order to determine whether the variable parameters and smoothing features of PeakCaller functioned as designed in reducing the incidence of type I and type II errors, we compared the rate of type I and type II errors in commercially available hiPSC-derived cortical neurons (d30 post differentiation) and dissociated mouse cortical neurons. In this instance, type I and type II errors were determined manually by the user. For interpretation of the results type I error is defined as the probability of rejecting the null hypotheses when the null hypotheses is statistically true (false positive). The type II error is incorrectly accepting the null hypotheses when the null hypothesis is statistically false (false negative) [22, 23]. Representative Ca2+ traces are presented to demonstrate what was called a type I and type II error with each experiment. Significance was determined by Chi square statistic, with an alpha of 0.05.
PeakCaller is a MATLAB-compatible script developed to allow users greater control over peak parameters and to apply data smoothing algorithms designed to reduce type I and type II errors in Ca2+ signaling studies in cultured neurons. This script and additional information can be found at www.hussmanautism.org/resources/software. While developed and applied to Ca2+ imaging in this study, PeakCaller can be used for any data set in which there is a need to detect the number and frequency of peak changes in the dependent variable over time. Other applications include financial data, solar emissions, and epidemiological modeling. In the instance of Ca2+ signaling, a “peak” represents the rapid increase of intracellular Ca2+ needed to trigger disparate cellular signaling cascades. At the cellular level, the raw data for PeakCaller can be collected in multiple ways (for a detailed review of Ca2+ signaling studies and Ca2+ indicators see: [24]). In most instances the raw data output from these various methods is similar: a time-dependent change in the intensity of the fluorescence signal within a defined ROI.
Whether performed on a traditional confocal microscope like the Zeiss Spinning Disk, or an automated high-throughput system like the ThermoFisher Array Scan, the user will specify a ROI within the recorded field (in the case of a neuron, typically a selected cell body or dendrite). The average fluorescence intensity of the pixels within the ROI will be recorded for each time point and can be exported according to the software used by the recording platform. PeakCaller is designed to automate the process of determining the number and frequency of Ca2+ signaling events (as estimated by temporal increases in the mean fluorescence per ROI) in an intuitive manner. In addition to analyzing Ca2+ signaling data, PeakCaller has an additional useful function particular to high-throughput systems like the Array Scan platform. PeakCaller will remove from the analysis incomplete data sets, which can arise when a cell body is dropped from the recording due to a significant decrease in the fluorescent signal measured during the automation process.
PeakCaller is easy to run and provides a simple way to visualize results, set appropriate analytics parameters, and record results in a concise and convenient format. When PeakCaller is started, only a single button labeled “Choose File” is active. Clicking this button opens a standard dialog box that prompts the user to pick a data file for analysis. For this manuscript, two imaging systems, the Zeiss Spinning Disk Confocal (Carl Zeiss AG, Oberkochen, Germany) and the ThermoFisher Array Scan (ThermoFisher Scientific Waltham, MA, USA) were used to generate the datasets used in the analysis; these datasets were saved as comma-delimited files (*.csv). In addition to being the two major platforms used for obtaining cell-based Ca2+ signaling, the Zeiss platform represents a low-to-medium throughput system, whereas the ThermoFisher system represents a high-throughput system. These two different systems produce data in two different formats. PeakCaller makes a quick check of the formatting to determine which type of data is present. (Examples of the different datasets are included on the website along with PeakCaller. Readers are encouraged to consult the Zeiss Zen Blue and Array Scan HCS Studio 2.0 software for detailed information of performing Ca2+ signaling experiments.)
Once a data set is successfully loaded, the software displays a button labeled “Find Peaks,” along with a panel that allows the user to fine-tune the qualities they require of peaks in their data. First, the user can choose to remove a background trend from the data, using the methods detailed in the section “PeakCaller–Smoothing Features.” Second, the user can define the characteristics that a peak should have: how far and how fast the data must rise and fall for a point to be characterized as a peak, as noted in the section “PeakCaller–Paramterizing a Peak.” These two key features are major improvements over the simpler PeakFinder MATLAB script, in that they allow the user more control over peak definition, and they adjust for global time-based changes in the intensity of the fluorescent signal that may occur during the course of an experiment, such as photobleaching.
Once the user has selected the parameters that best represent their data, they click the “Find Peaks” button and the results are displayed visually on two graphs–the upper graph shows the original data and the defined long-term trend, while the lower graph shows the de-trended data. Each has all identified peaks marked with small green circles. A scroll bar below the graphs allows the user to quickly see the results for all regions of interest included in the data file.
The peak data is also displayed in the MATLAB command window, and the user can choose to extract the lower graph (for easy manipulation and printing) or save the peak data to a file. For each region of interest, the software displays the number of events (peaks), the intervals between events, and the average frequency implied by those intervals. It also provides the average frequency of events across all ROIs, the mean number of events per ROI, and the standard deviation of the number of events per ROI.
PeakCaller: smoothing features
PeakCaller offers several tunable options for muting the effects of an underlying trend in a time series. Minimizing the influence of such trends can be important when there are global changes in background fluorescence due to bleaching or changes in focus that cause the measured baseline fluorescence to become nonlinear. PeakCaller approximates the underlying trend by generating a smoothed version of the time series, which emphasizes the long-term movements in the time series and deemphasizes the quick peaks and background fluctuations. PeakCaller includes four options for generating this smoothed series: the first three choices might be considered more traditional convolution-based smoothers, while the fourth has a more topological flavor. In any case, the resulting smoothed version of the time series serves as a rough approximation of the underlying trend, and PeakCaller controls for this trend simply by dividing the original time series by the smoothed profile. The four options for generating this smoothed background trend line are described here.
Finite difference diffusion
In physical systems, the process of diffusion describes the collective motion of particles. Real-world examples of diffusion include things like the movement of milk that has been poured into a cup of coffee or the spread of engine exhaust from the tailpipe of a car. Over time, particles move from areas of high-concentration to low, and everything becomes more and more uniform. In PeakCaller, finite difference diffusion numerically approximates this process—treating the original time series much like an initial measure of the spatial concentration of engine exhaust—and numerically approximating its diffusion and spread. Small values of the parameter trend smoothness correspond to short diffusion times (less smoothing), whereas larger values correspond to longer diffusion times (more smoothing). Figure 1 shows a representative recording from a hiPSC-derived neuron treated with Fluo-4. In this case, the smoothed profile shown was produced via finite difference diffusion. Additionally, the figure includes an overlay showing how the values of the original profile are weighted in order to generate the smoothed profile.
Fig. 1.
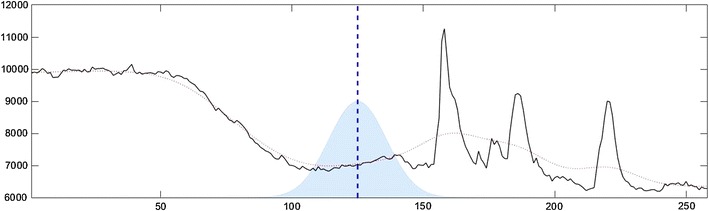
This figure illustrates the weight function and smoothing corresponding to finite difference diffusion, with the trend smoothness parameter set to 80. Time is on the horizontal axis (in seconds) and the fluorescence intensity of the Fluo-4 emission is on the vertical axis (arbitrary units). For the original Ca2+ recording (in black), the smoothing is given by the dotted red curve. To visualize the smoothing calculation, at the dashed blue line, the smoothed value is computed by weighting the values of the original profile proportionally with the weight function shaded in light blue. Notice that this weight function is approximately Gaussian
Exponential moving average (1-sided)
Moving averages provide an elementary technique for smoothing time series, and are often used in finance to highlight long-term trends. For any point in the data set, the exponential moving average depends only upon the data up to that point, with the weighting of the older data falling off exponentially. In PeakCaller, small values of the parameter trend smoothness correspond to fast-decaying exponentials (less smoothing), whereas larger values correspond to longer decay times (more smoothing). Figure 2 shows a representative recording from a hiPSC-derived neuron treated with Fluo-4. The smoothed profile was produced using an exponential moving average. Additionally, the figure includes an overlay demonstrating how the values of the original profile are weighted in order to generate the smoothed profile.
Fig. 2.
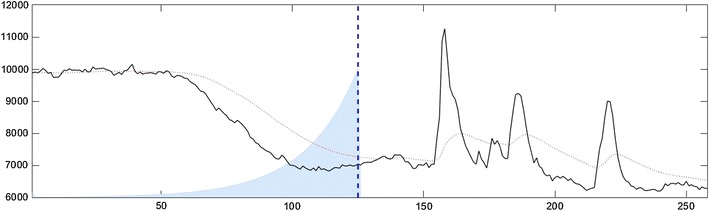
Example of the weight function and smoothing corresponding to a 1-sided exponential moving average with the trend smoothness parameter set to 40. For the original Ca2+ recording (in black), the smoothing is given by the dotted red curve. To visualize the smoothing calculation, at the dashed blue line, the smoothed value is computed by weighting the values of the original profile proportionally with the weight function shaded in light blue. Note especially that the smoothed value at any point depends only on the portion of the original profile to the left of that point
In practice, this method is particularly useful for data sets with a higher degree of changes in background fluorescence, due to conditions such as overgrown (multi-layered) cell cultures, cultures with an excess degree of dead cells, or weak Ca2+ signals that may be hard to distinguish from background. This method is recommended when changes in culture conditions over time are expected due to pharmacological intervention, which could affect the characteristics of future peaks.
Exponential moving average (2-sided)
Exponential moving averages are commonly used with live data, where only the past history is known; an estimate of the current trend cannot make use of future data that is not yet in evidence. In the current context however, we already have the entire recording of Ca2+ transients from a given time window, and thus can approximate the trend at any point by using measurements taken both before and after that particular moment in time. One option is to simply average a backward-looking exponential moving average (looking back to the beginning of the recording) with a forward-looking exponential moving average (looking forward to the end of the recording). This method might be recommended over the classic exponential moving average (1-sided) when culture conditions remain static.
Figure 3 shows a representative recording from a hiPSC-derived neuron treated with Fluo-4 and the smoothed profile produced using the 2-sided exponential moving average. Additionally, the figure includes an overlay showing how the values of the original profile are weighted in order to generate the smoothed profile. Note that these weights differ from the other two-sided weights given via finite-difference diffusion in Fig. 1.
Fig. 3.
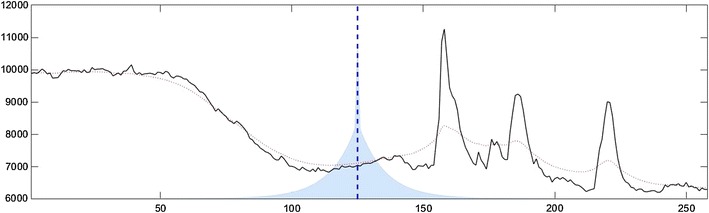
Graphical representation of the weight function and smoothing corresponding to a 2-sided exponential moving average with the trend smoothness parameter set to 20. For the original Ca2+ profile (in black), the smoothing is given by the dotted red curve. To visualize the smoothing calculation, at the dashed blue line, the smoothed value is computed by weighting the values of the original profile proportionally with the weight function shaded in light blue. Notice especially the double-weight given to the value at the blue line, as it is the only point in common to both the forward- and backward-looking moving averages
Convex envelope
This is a purely geometric measure that defines the trend as the portion of convex envelope underneath the graph of the time series. Intuitively, one can picture an elastic band running under the graph and tied to both end points. When that band is snapped tight along the bottom, the resulting convex envelope can be used as a measure of the trend. In PeakCaller, the parameter trend smoothness plays no role in defining the trend in this case, as the smoothing is defined in a parameter-free way by an advanced game of “connect the dots”. The smoothing and the resulting de-trended profile are both displayed in Fig. 4.
Fig. 4.
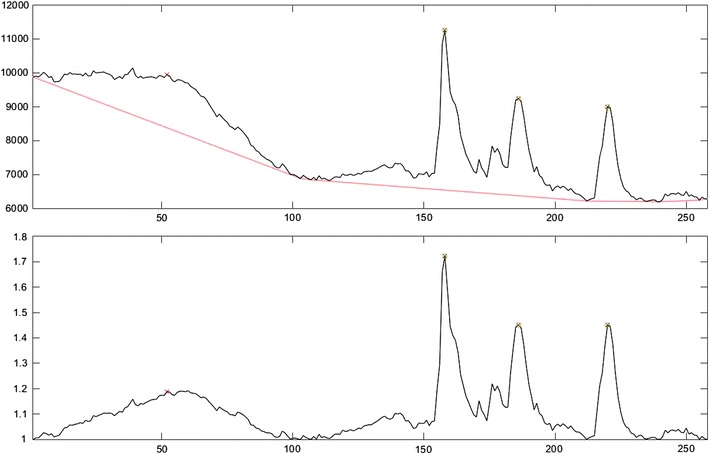
Illustration of smoothing using the convex envelope and shows the actual graphics provided by PeakCaller. In the upper graph, for the original Ca2+ recording (in black), the smoothing using a convex envelope is given by the dotted red curve. The lower graph shows the de-trended data series, which is simply the quotient of the original data divided by the smoothed data. Note in this case that the de-trended data “bounces off” of a minimum value of one exactly at the corners where the convex envelope touches the original profile. Peaks identified by PeakCaller are marked with a green circle, whereas peaks identified by PeakFinder are marked with a red ‘X’
No trend
In instances where the original data is linear in nature and no change to culture conditions is suspected, there is an option to leave the raw data untransformed. When no trend is chosen, the mean of the original profile is used as a trivial smoothing. The de-trended profile displayed by PeakCaller will then have exactly the same shape as the original profile, but scaled so that the mean is 1.
PeakCaller: parameterizing a peak
PeakCaller characterizes peaks using a clear and straightforward set of criteria. In order for a point to be called a peak, the de-trended data must rise a significant amount in a well-defined window just before the candidate point. The user can define the size of the rise that they deem significant (by setting required rise (%) in PeakCaller) as well as the maximum length of the window allowed for the rise to occur (max look-back (pts) in PeakCaller). The actual look-back window (the time interval in which the florescent must rise in order to be called a peak) will be shortened appropriately if either (a) the max look-back goes beyond a previously identified peak or (b) the max look-back goes beyond the beginning of the data.
In complementary fashion, for a point to be a peak, the de-trended data must fall a significant amount in a well-defined window following the candidate point. The user can define the size of the fall that they deem significant (by setting required fall (%) in PeakCaller) and the maximum length of the window allowed for the fall to occur (max look-ahead (pts) in PeakCaller). The actual look-ahead window will be shortened appropriately if either (a) the max look-ahead goes beyond the end of the data or (b) the max look-ahead includes points that are higher than the candidate.
A diagram showing the four quantities that a user may use to define a peak is given in Fig. 5. Although many users may choose matching values for rise and fall, as well as look-back and look-ahead, PeakCaller uncouples these values to allow the user some versatility. In many applications, signals are not perfectly symmetric–with depolarization and repolarization happening on different timescales–and the user now has the freedom to vary these parameters as they deem appropriate.
Fig. 5.
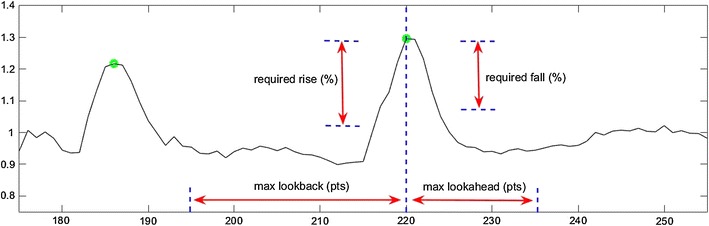
This figure details the parameters used by PeakCaller to characterize a peak. For a point to be classified as a peak there must be a significant rise over a designated interval before the point, and a significant fall within a designated interval after the point
PeakCaller’s GUI allows users without MATLAB coding experience to modify peak parameters to best interpret their data. The features described above coupled with the GUI are intended to be an improvement over the original PeakFinder algorithm. PeakFinder classifies peaks according to an approximation for the area of candidate peak formations. This scoring for peaks rewards the wide candidate peak formations in a way that is not always appropriate in the search for quick localized spikes. Also, in developing cortical neuron cultures, Ca2+ peaks may not have a time scale as well developed as that in dissociated mouse cortical neurons. The ability to set culture-specific look-back windows is intended to allow the user to modify the peak calling script to suit the developmental state of their cultures. The PeakFinder script was not originally designed to process Ca2+ signaling data and thus is not sensitive to the affinity and specificity of Ca2+ indicators. By allowing for the user to set custom look-back windows, PeakCaller is able to prevent type II errors and the rejection of a smaller peak in close proximity to a larger peak. Once the parameters have been set, PeakCaller automates the peak calling process for all regions of interest in the comma delimited (.csv) file, greatly increasing the efficiency of peak calling over manual peak calling.
Graphically, PeakCaller will denote a peak by placing a green circle around the point in both the original and the de-trended data. Should the user wish to compare their selected peak parameters and smoothing function to de-trended data using the “outstanding area” method of peak calling found in PeakFinder, the user can select the “show old peaks” box, which will display the original PeakFinder peaks using a red ‘X’.
PeakCaller: visualization and connectivity
In order to have a large scale visual representation of the traces in the context of the entire experiment, the view traces button can be used. Pressing the button labelled View Traces allows the user to see all of the fluorescence traces in a single figure, with the identified peaks highlighted in a different color. An example of this in a high content assay run across ~ 500 human iPSC-derived neurons expressing GCamp6 using the ThermoFisher ArrayScan and ~ 200 neurons treated with Fluo-4 and imaged at 10 Hz is presented in (Fig. 6). After visualization, the histograms button opens up two histograms: one summarizing the heights of the many peaks and the other the rise and fall times. These can be used to determine general characteristics of the peaks and help to get a population-based measurement of the transient behavior. To develop a general overview of the temporal clustering of Ca2+ transients, the end user can generate a raster plot. Pressing the button labelled Raster Plot allows the user to see a raster plot of all of the identified peaks in a single figure. This can be used as a way to easily visualize clusters of related activity overall in the data set.
Fig. 6.
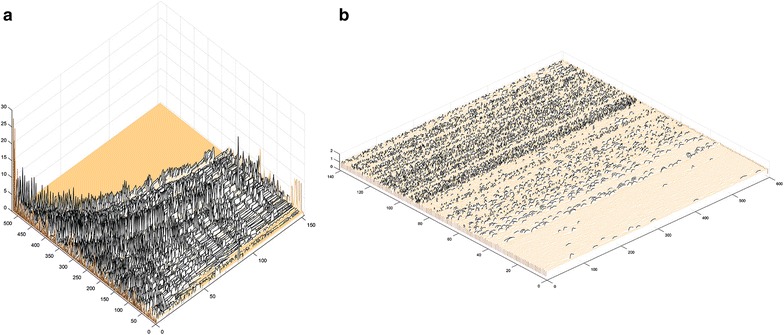
Multiple traces and ROIs can be visualized using the Peak Caller. The top dark parts of the trace are the identified peaks for each time series. This visualization can be rotated to examine the activity across an entire data set. a Ca2+ transients recorded from ~ 500 human iPSC-derived neurons transfected with the Ca2+ indicator GCamp6 recording using the ThermoFisher ArrayScan system. b Ca2+ transients recorded from ~ 200 human iPSC-derived neurons treated with the Ca2+ indicator Fluo-4 imaged at 10 Hz (x-axis represents time in seconds, y-axis represents number of ROIs and z-axis represents amplitude as ΔF/F)
To examine whether there is a functional relationship between ROIs, possibly determinant of neuronal connectivity, there are two types of measurements that PeakCaller offers. The first is by clicking the button labelled Correlations. PeakCaller then makes a run through all of the traces, looking pairwise for evidence of a relationship between each individual trace and the temporal shifts of the others. The results are then displayed graphically in a heat map. Note especially that the pairwise correlations computed here are based upon the original traces and not upon the software’s identification of individual peaks.
Alternatively, a synchrony index can be constructed and calculated from the full set of ROIs. This index is computed by clicking the button labelled Synchronicity. In this case, the original traces are replaced with a bare-bones version that takes the value one near each peak (above the half-maximum value of the peak) and zero elsewhere. A generalized autocorrelation function is then produced for each of these traces, and a synchronization index is computed pairwise based on these autocorrelation functions. This method is similar to the one detailed in [25]. Because of the construction of the generalized autocorrelation functions, the pairwise indices should be the largest when the two ROIs have peaks that are similarly spaced, even if one series lags the other significantly. These pairwise values are then displayed graphically in a heat map. A second heat map is also displayed, with the ROIs rearranged in an attempt to group them into identifiable clusters. At the top of both heat maps, a “global” or “mean” index is displayed, representing the average index across all of the different pairs. This global/mean index should be closest to one if a large amount of pairwise synchrony is detected across all of the ROIs in the data set. This synchronization index, in combination with the correlations and the raster plot, can give the end user insight into whether the set of ROIs in the data set are functionally connected (Fig. 7).
Fig. 7.
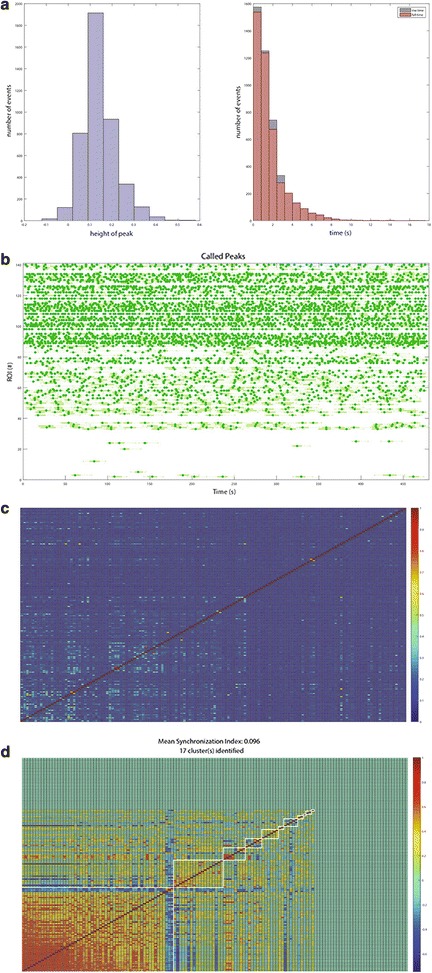
Typical functional output for a high-content Ca2+ imaging experiment. a Histograms, b raster plots, c correlation, and d synchronization indexes can be calculated for large numbers of cells to determine group characteristics of calcium transients in a high-throughput screen. The heat maps and synchronization index can give the end user a measure of the functional connectivity in a given dataset. In this dataset the Ca2+ transients are not functionally connected, as evidenced by no bursting patterns in the raster plot, low autocorrelations in the heat map, and a synchronicity index of 0.096. This is to be expected, as these transients were recorded from neural progenitors at day 21 derived from human iPSCs
Results
In this study we use Fluo-4, a [Ca2+] indicator, and analyze Ca2+ signaling in commercially available hiPSC-derived cortical neurons and dissociated mouse cortical neuron cultures. Raw output files from the Zeiss Spinning Disk and ThermoFisher ArrayScan platforms were analyzed using PeakCaller. PeakCaller efficiently and accurately imported both data files and calculated the number of peaks per ROI (cell body) and peak intervals as shown in Fig. 8. Specific parameters were chosen for each file and analyzed accordingly. PeakCaller reduced the incidence of type I errors in day 30 cultured iPSC derived cortical neurons (p < 0.05) (Fig. 9) in three separate recordings. The incidence of type II errors was unaffected by the use of the PeakCaller script. The representative trace provided in Fig. 9 demonstrates how PeakFinder overestimates the number of peaks when presented with a signal that represents a high fluctuation in the background fluorescence. The PeakFinder algorithm uses an a priori approach that assumes that a peak is present, as it denotes a peak based on the highest point within a set window, and thus will always produce false positives in an instance where there is a background of fluorescence.
Fig. 8.
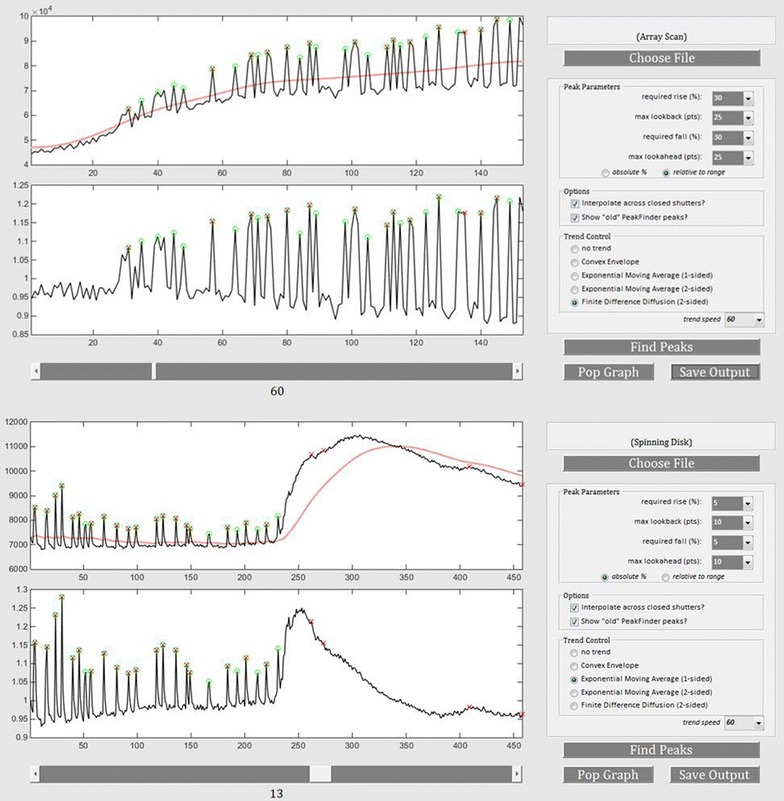
Analysis of Zeiss Spinning Disk and ThermoFisher Array Scan Ca2+ imaging data output by PeakCaller. The vertical axis represents Fluo-4 fluorescence emission intensity (arbitrary units) and the horizontal axis is time in seconds. Green circles represent PeakCaller identified peaks. Red Xs represent PeakFinder identified peaks. Parameters were chosen to best represent data generated on both platforms
Fig. 9.
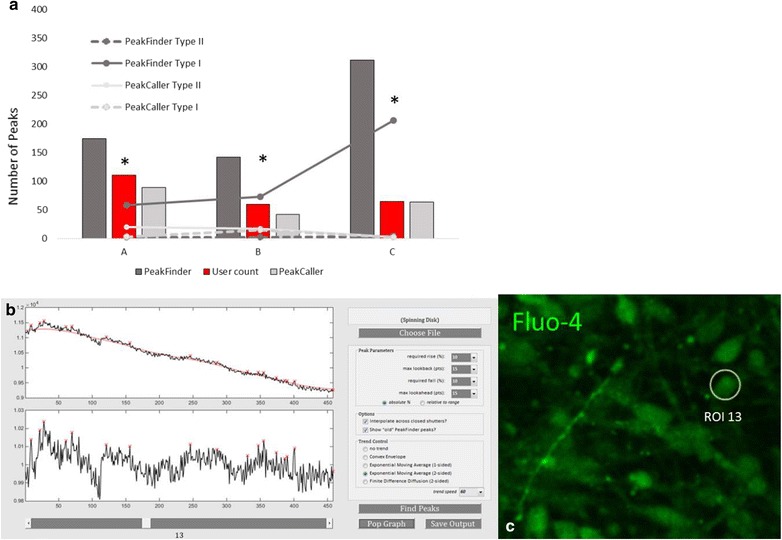
PeakCaller decreases the incidence of type I errors in cultured hiPSC-derived cortical neurons (*p < 0.05). a No difference in incidence of type II errors was found between PeakCaller and PeakFinder under these culture conditions. b Representative trace shows over estimation of peak incidence and frequency in PeakFinder (red X) script. c Designated ROI for the recording in this representative trace
PeakCaller has several advantages over PeakFinder in these instances because the user can set a minimum rise and fall within a window (needed to denote a “peak”), as well as the width of the window to accurately represent the fluctuation of the data. The ability to accurately represent the frequency of Ca2+ signaling in developing neurons by reducing the instance of type I and type II errors is crucial to the identification of cellular phenotypes associated with neurological conditions and experimental manipulation. Once phenotypes associated with aberrant Ca2+ signaling have been identified, the program can be used to accurately measure the efficacy of pharmacological interventions. In Fig. 9, PeakCaller accurately identified 0 peaks in the ROI, while PeakFinder identified 17 false positive peaks.
Pharmacological manipulation is an important tool used to interrogate neuronal function. Picrotoxin (PTX) is a gamma-aminobutyric acid-alpha (GABAA) receptor antagonist that can induce rapid firing of action potentials and potentially glutamatergic excitotoxicity by significantly decreasing the activity of inhibition in neuronal circuits [26, 27]. After the application of PTX to cultured neurons, cells will enter a state of heightened excitability followed by loss of excitation after exhaustion of glutamate stores. This change in excitation and inhibition can be measured by looking at the activity of Ca2+ in neurons [27]. We exploited this function of PTX in two model systems in order to test the functionality of the PeakCaller script and compare it to PeakFinder.
To do this, we exposed d30 hiPSC-derived cortical neurons to 50 μM PTX with a 1 min wash in phase and 40 min recording (four 10 min recordings). The raw data was analyzed using both PeakCaller and PeakFinder. PeakCaller accurately identified windows of high Ca2+ activity in neurons, while PeakFinder consistently over-estimated the number of peaks and number of active cells (Fig. 10). In this context, PeakFinder becomes less accurate and more vulnerable to type I error as the number of true peaks decreases relative to background fluorescence.
Fig. 10.
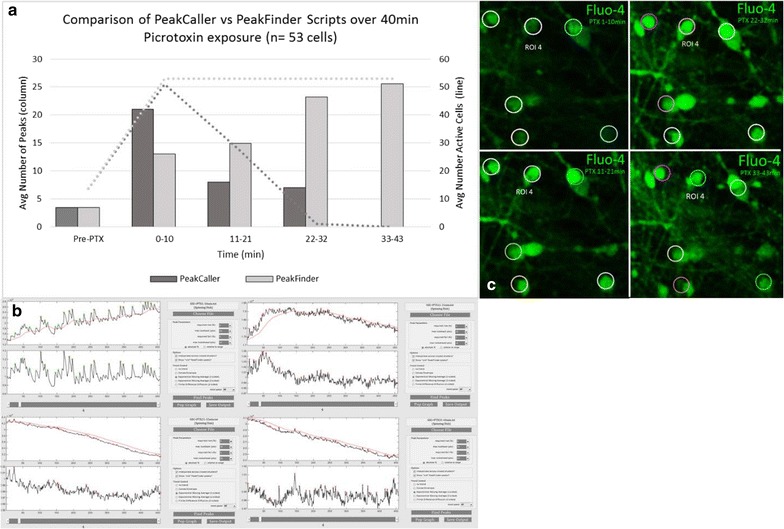
a Reduction of type I errors by PeakCaller after application of PTX to induce excitation. Prior to PTX exposure both PeakFinder and PeakCaller correctly identified the same number of peaks per active cell. After PTX exposure (50 μM), PeakCaller correctly modeled the state of induced excitation and excitotoxicity in the hiPSC derived cortical neurons, while PeakFinder initially under estimated the number of peaks and then over estimated the number or Ca2+ peaks. b Representative traces of ROI 4 are provided to show the transient increase in Ca2+ signaling after application of PTX. c Designated ROIs across all recordings for this representative experiment
The incidence of type I and type II errors in PeakCaller. PeakFinder scripts were also compared in the analysis of dissociated mouse cortical neurons. While a direct comparison of Ca2+ signaling in mouse and hiPSC-derived cortical neurons has not yet been published, it would be expected that these two cultures may have different Ca2+-transient characteristics.
In four separate recordings, PeakCaller reduced the incidence of type II errors in three of the recordings. There was no difference in the incidence of type II errors in the fourth recording (Fig. 11). PeakCaller also indicated a significant (p < 0.05) reduction in type I error in recording A, but no difference in type I errors in the other recordings. In the representative trace in Fig. 4, PeakFinder identified only two peaks, whereas the user and PeakCaller identified 26 and 28 peaks, respectively. In this instance PeakFinder underestimated the number of peaks present for the same reason it over-estimated the number of peaks in the previous experiments. PeakFinder selected the two highest points in the data set as peaks and disregarded true peaks that fell below these higher points, due to PeakFinder’s inability to normalize the data set to changes in fluorescence. PeakCaller’s smoothing functions can be set to account for global changes in fluorescence and can be set with narrower look-back and look-ahead windows to properly identify the majority of peaks in a given ROI.
Fig. 11.
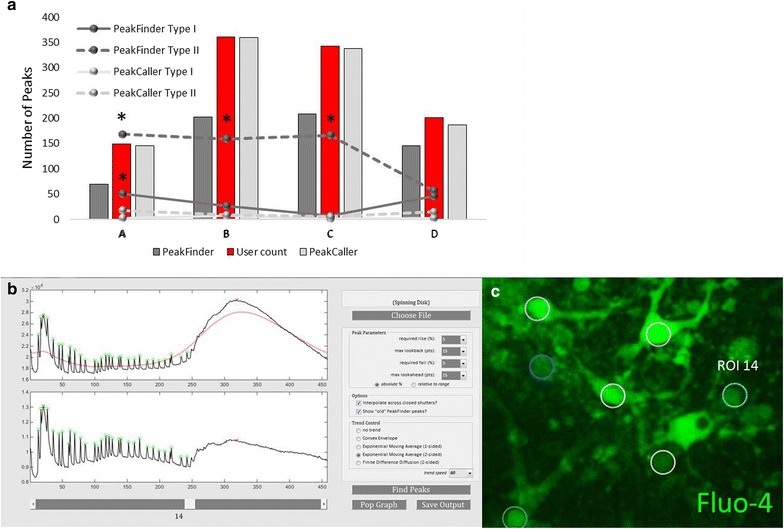
a Reduction of type II error in dissociated mouse cortical neurons. PeakCaller demonstrated the ability to reduce the occurrence of type II errors (p < 0.05) in three of four Ca2+ recordings. No difference in incidence of type II error was found between the two scripts for the fourth recording. PeakCaller also showed a significant (p < 0.05) reduction in type I error in recording A. b Representative traces below show PeakCaller peaks in green circles and PeakFinder peaks as red Xs. c Designated ROIs for this representative experiment
Conclusions
Here we have demonstrated that PeakCaller can reduce the incidence of both type I and type II errors in hiPSC-derived cortical neurons, dissociated mouse cortical neurons, and cells in a state of induced neuronal excitation. PeakCaller’s advantages stem from two main features: (1) smoothing functions that can normalize changes in background fluorescence, and (2) settable parameters for minimum peak height and look-back/ahead windows. The script does not achieve perfect accuracy in its designation of Ca2+ peaks, but it significantly outperforms PeakFinder with respect to reduction in both types of statistical error. The fact that PeakCaller can reduce different types of error produced by different cell line characteristics and culture conditions is important to the study of neurodevelopmental, neuropsychiatric, and neurodegenerative diseases, particularly in the context of high-throughput screening. It is foreseeable that a disease condition may produce a phenotype that is more vulnerable to the significant type I or type II error bias introduced by the use of PeakFinder, and as such may confound any data analyzed using this algorithm.
PeakCaller allows the user freedom to designate peak parameters and smoothing functions to most accurately represent the raw data within a GUI that is accessible to users without extensive MATLAB or coding experience. Designed specifically to address the conditions/limitations of Ca2+ signaling in cultured neurons, PeakCaller is a powerful new tool in the hands of the neurophysiologist.
Authors’ contributions
EA, RJ, MK, MWN, performed experiments. RJ performed the programming. MWN conceived the idea. EA analyzed the data. All authors read and approved the final manuscript.
Acknowledgements
We thank Elizabeth Benevides for proofreading the article and Drs. John Hussman and Gene Blatt as well as members of the Hussman Institute for Autism for helpful discussions and comments. Thank you to Drs. Tom Blanpied and Joe Mauban for help with confocal microscopy.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Project name: PeakCaller.
Project home page: www.hussmanautism.org/resources/software.
Operating System: Platform independent.
Programming Language: MATLAB.
Other requirements: Matlab software R2104a or higher.
License: None.
Any restrictions to use by non-academics: None.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Animal work was reviewed and approved by institutional IACUC committee (IACUC protocol at the Hussman Institute for Autism: #06012015B; IACUC protocol at University of Maryland at Baltimore: #0515016).
Funding
This work was supported by a pilot grant from The Hussman Foundation to MWN and YCL. The Hussman Foundation played no role in the writing of the manuscript, design of the study, collection, analysis or interpretation of the results.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Ca2+
calcium
- hiPSC
human induced pluripotent stem cell
- ROI
region of interest
- PTX
picrotoxin
- GABAA
gamma-aminobutyric acid-alpha receptor
Footnotes
Elena Artimovich and Russell K. Jackson contributed equally
Contributor Information
Elena Artimovich, Email: geneticsbabe@gmail.com.
Russell K. Jackson, Email: rjackson@hussmanfunds.com
Michaela B. C. Kilander, Email: mkilander@hussmanautism.org
Yu-Chih Lin, Email: yclin@hussmanautism.org.
Michael W. Nestor, Email: mnestor@hussmanautism.org
References
- 1.Sutherland DJ, Goodhill GJ. The interdependent roles of Ca(2+) and cAMP in axon guidance. Dev Neurobiol. 2015;75:402–410. doi: 10.1002/dneu.22144. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg SS, Spitzer NC. Calcium signaling in neuronal development. Cold Spring Harb Perspect Biol. 2011;3:a004259. doi: 10.1101/cshperspect.a004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans RM, Zamponi GW. Presynaptic Ca2+ channels–integration centers for neuronal signaling pathways. Trends Neurosci. 2006;29:617–624. doi: 10.1016/j.tins.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Zucker RS. Calcium- and activity-dependent synaptic plasticity. Curr Opin Neurobiol. 1999;9:305–313. doi: 10.1016/S0959-4388(99)80045-2. [DOI] [PubMed] [Google Scholar]
- 5.Demuro A, Parker I, Stutzmann GE. Calcium signaling and amyloid toxicity in Alzheimer disease. J Biol Chem. 2010;285:12463–12468. doi: 10.1074/jbc.R109.080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmieri L, Papaleo V, Porcelli V, Scarcia P, Gaita L, Sacco R, Hager J, Rousseau F, Curatolo P, Manzi B, Militerni R, Bravaccio C, Trillo S, Schneider C, Melmed R, Elia M, Lenti C, Saccani M, Pascucci T, Puglisi-Allegra S, Reichelt KL, Persico AM. Altered calcium homeostasis in autism-spectrum disorders: evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Mol Psychiatry. 2010;15:38–52. doi: 10.1038/mp.2008.63. [DOI] [PubMed] [Google Scholar]
- 7.Wen Y, Alshikho MJ, Herbert MR. Pathway network analyses for autism reveal multisystem involvement, major overlaps with other diseases and convergence upon MAPK and calcium signaling. PLoS ONE. 2016;11:e0153329. doi: 10.1371/journal.pone.0153329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nestor MW, Phillips AW, Artimovich E, Nestor JE, Hussman JP, Blatt GJ. Human inducible pluripotent stem cells and autism spectrum disorder: emerging technologies. Autism Res. 2016;9:513–535. doi: 10.1002/aur.1570. [DOI] [PubMed] [Google Scholar]
- 9.Berridge MJ. Dysregulation of neural calcium signaling in Alzheimer disease, bipolar disorder and schizophrenia. Prion. 2013;7:2–13. doi: 10.4161/pri.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berridge MJ. Neuronal calcium signaling. Neuron. 1998;21:13–26. doi: 10.1016/S0896-6273(00)80510-3. [DOI] [PubMed] [Google Scholar]
- 11.Brini M, Cali T, Ottolini D, Carafoli E. Neuronal calcium signaling: function and dysfunction. Cell Mol Life Sci. 2014;71:2787–2814. doi: 10.1007/s00018-013-1550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Illes S, Theiss S, Hartung HP, Siebler M, Dihné M. Niche-dependent development of functional neuronal networks from embryonic stem cell-derived neural populations. BMC Neurosci. 2009;10:93. doi: 10.1186/1471-2202-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel TP, Man K, Firestein BL, Meaney DF. Automated quantification of neuronal networks and single-cell calcium dynamics using calcium imaging. J Neurosci Methods. 2015;243:26–38. doi: 10.1016/j.jneumeth.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackay L, Mikolajewicz N, Komarova SV, Khadra A. Systematic characterization of dynamic parameters of intracellular calcium signals. Front Physiol. 2016;7:525. doi:10.3389/fphys.2016.00525. [DOI] [PMC free article] [PubMed]
- 17.Jang MJ, Nam Y. NeuroCa: integrated framework for systematic analysis of spatiotemporal neuronal activity patterns from large-scale optical recording data. Neurophotonics. 2015;2:035003. doi: 10.1117/1.NPh.2.3.035003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong LC, Lu B, Tan KW, Fivaz M. Fully-automated image processing software to analyze calcium traces in populations of single cells. Cell Calcium. 2010;48:270–274. doi: 10.1016/j.ceca.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Kaifosh P, Zaremba J, Danielson N, Losonczy A. SIMA: Python software for analysis of dynamic fluorescence imaging data. Front Neuroinform. 2014;27(8):77. doi: 10.3389/fninf.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaifosh P, Lovett-Barron M, Turi GF, Reardon TR, Losonczy A. Septo-hippocampal GABAergic signaling across multiple modalities in awake mice. Nat Neurosci. 2013;16(9):1182–1184. doi: 10.1038/nn.3482. [DOI] [PubMed] [Google Scholar]
- 21.Beaudoin GM, 3rd, Lee SH, Singh D, Yuan Y, Ng YG, Reichardt LF, et al. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat Protoc. 2012;7(9):1741–1754. doi: 10.1038/nprot.2012.099. [DOI] [PubMed] [Google Scholar]
- 22.Sheskin DJ. Handbook of parametric and nonparametric statistical procedures. Boca Raton: Chapman and Hall; 2011. [Google Scholar]
- 23.Neyman J, Pearson ES, Yule GU. The testing of statistical hypotheses in relation to probabilities a priori. Math Proc Camb Philos Soc. 1933;29:492. doi: 10.1017/S030500410001152X. [DOI] [Google Scholar]
- 24.Grienberger C, Konnerth A. Imaging calcium in neurons. Neuron. 2012;73:862–885. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 25.Holliday J, Adams RJ, Sejnowski TJ, Spitzer NC. Calcium-induced release of calcium regulates differentiation of cultured spinal neurons. Neuron. 1991;7:787–796. doi: 10.1016/0896-6273(91)90281-4. [DOI] [PubMed] [Google Scholar]
- 26.Kidd FL, Isaac JT. Glutamate transport blockade has a differential effect on AMPA and NMDA receptor-mediated synaptic transmission in the developing barrel cortex. Neuropharmacology. 2000;39:725–732. doi: 10.1016/S0028-3908(99)00270-1. [DOI] [PubMed] [Google Scholar]
- 27.Eisenman LN, Kress G, Zorumski CF, Mennerick S. A spontaneous tonic chloride conductance in solitary glutamatergic hippocampal neurons. Brain Res. 2006;1118:66–74. doi: 10.1016/j.brainres.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Project name: PeakCaller.
Project home page: www.hussmanautism.org/resources/software.
Operating System: Platform independent.
Programming Language: MATLAB.
Other requirements: Matlab software R2104a or higher.
License: None.
Any restrictions to use by non-academics: None.


