Fig. 4.
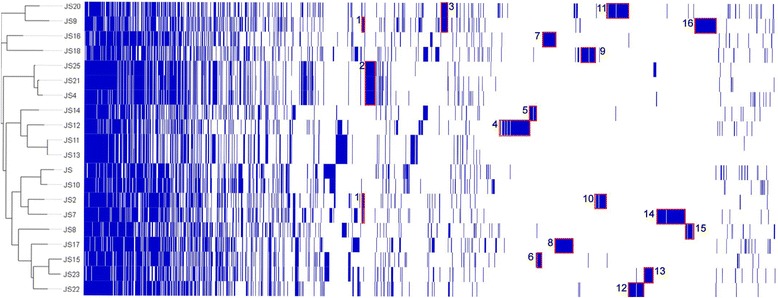
Map of accessory genome alignments generated by Roary. Gene clusters unique to individual strains are marked in red and numbered. 1) Genomic island with genes coding for the CASCADE-like CRISPR-Cas systems in strains JS2, JS7 and JS9; 2) Genomic island unique to strains JS4, JS21 and JS25. Genes located on this island include transposase genes with 96–98% sequence identity to those from Corynebacterium urealyticum DSM 7111 and a gene coding for an additional Cobyrinic acid A,C-diamide synthase; 3) Heat shock island unique to strains JS9 and JS20. Genes on the island include an 18-kDa heat shock protein, DnaK, GrpE, CbpM, ClpB and others; Features 4–16 are gene clusters unique to their respective strains. These include complete prophages (8, 12, 13, and 14), phage remnants (6, 8, 9 and 12), predicted genomic islands with genes encoding various functions: resistance to heavy metals (7), possible antibiotic resistance (15), genetic loci with genes coding for restriction and modification systems (7, 11, 12 and 14), and the pilus locus (9). Unique gene clusters 4, 9, 11, 14 and 16, despite sequence differences share structural similarities, including the presence of genes coding for Single-stranded DNA-binding protein, TraM recognition site of TraD and TraG, AAA-like domain protein (VirB4-like), Multifunctional conjugation protein TraI (TrwC or TraA relaxase), Type IV secretory system Conjugative DNA transfer (TraG-like), ParB-like nuclease domain protein, Bifunctional DNA primase/polymerase and Murein DD-endopeptidase MepM. The presence of TraA, TraG and VIrB4 are indicative of Integrative and Conjugative Elements (ICEs) type T4SS. The majority of the unique gene clusters have regions with a high degree of sequence identity to other Actinobacteria, including Propionibacterium acidipropionici, Corynebacterium falsenii, Cutibacterium avidum and Microbacterium sp. Details can be viewed in the Additional tables of the respective strains in the column “Note”
