Table 5.
Methylation motifs and the responsible methylases identified in P. freudenreichii
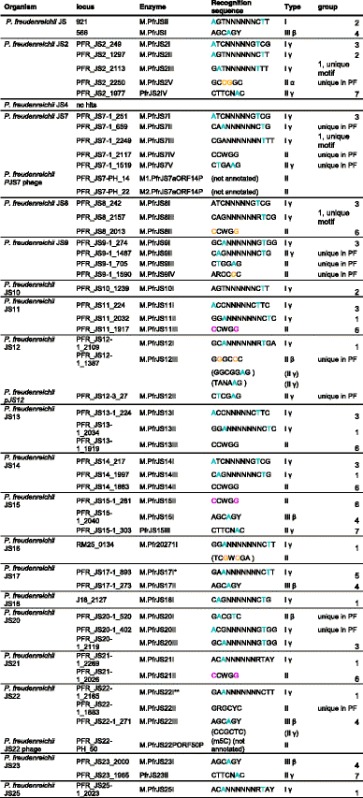
Recognition sequences in parentheses indicates the methylase responsible cannot be assigned unambiguously. Modified bases are marked in colour; “not annotated” indicates there is no locus tag as the gene is not annotated in the GenBank file. Color coding for the methylation types Blue: 6 mA; Orange: 4mC; Purple: 5mC
*It is possible that it is really M.PfrJS17ORF2252P that is the active methylase in this organism. M.PfrJS17ORF2252P shares 99% aa sequence identity with M.PfrJS22I, but its corresponding S protein is truncated
**It is possible that it is really M.PfrJS22ORF850P that is the active methylase in this organism. M.PfrJS22ORF850P is 100% identical to M.PfrJS17I, as are their corresponding S proteins while the R proteins differ by one amino acid (475 Ala:Gly)
