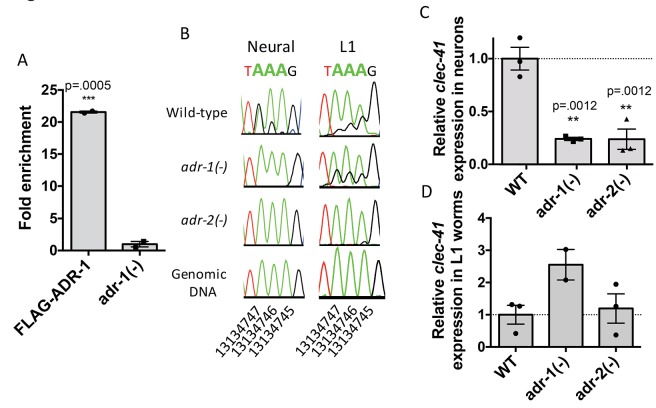
Figure 4. Neural A-to-I RNA editing and expression of clec-41 is regulated by ADR-1 and ADR-2.
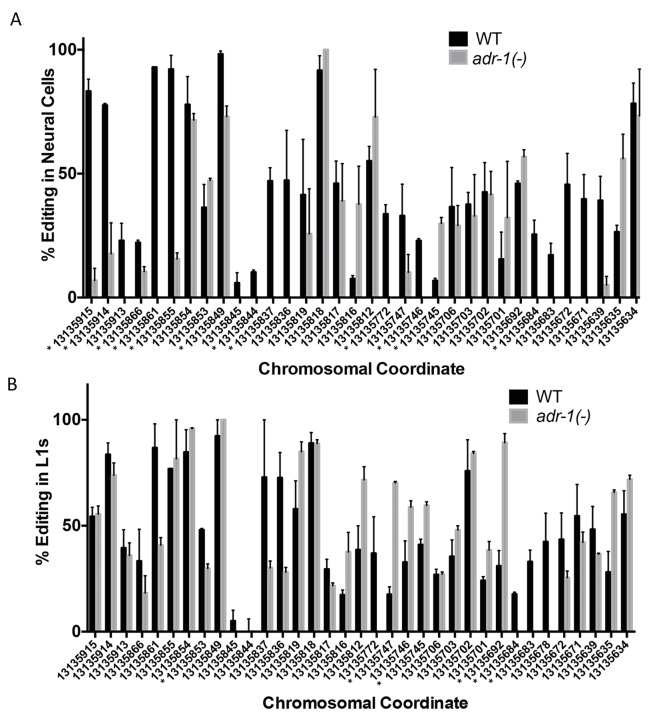
(A) Lysates from adr-1(-) worms and worms expressing FLAG-ADR-1 were subjected to a FLAG immunoprecipitation (IP). qRT-PCR was performed on both RNA from the input lysates as well as the IP samples. The levels of clec-41 in the IP samples was divided by the level of clec-41 in the lysate and the fold enrichment of this ratio for FLAG-ADR-1 normalized to the negative control adr-1(-) was determined. The average of two independent biological replicates is plotted with error bars representing the SEM. Student’s t-test, ***p<0.001. (B) Sanger sequencing chromatograms of clec-41 genomic DNA or cDNA amplified from the indicated strains. The chromosomal coordinates (ce11) for the edited adenosines in the wild-type cDNA are indicated below the chromatograms, representative from three (Neural) or 2 (L1) independent biological replicates (Quantification of all editing sites can be seen in Figure 4—figure supplement 1) RNA isolated from neural cells (C) or L1 whole worms (D) for the indicated strains was subjected to reverse transcription and qRT-PCR to determine levels of clec-41 from three independent biological replicates. The average expression of clec-41 relative to the house-keeping gene, gpd-3 were normalized to WT and plotted with SEM. One-way ANOVA followed by Dunnett’s Multiple Comparisons Correction, **p<0.01.