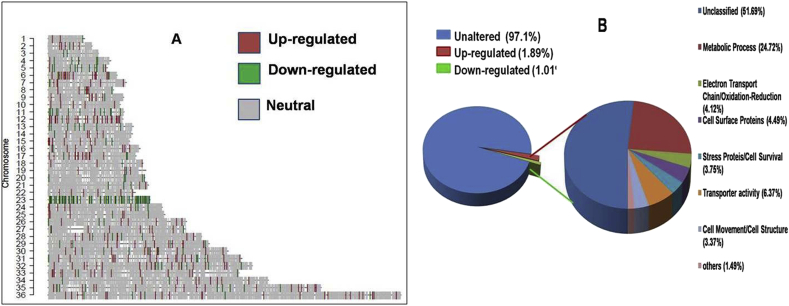
Fig. 2.
A. Comparative gene expression of PMM-S and PMM-R parasite on chromosomal map. Chromosome map for expression data was generated using Custom R program. Red and green lines indicate up regulated and down-regulated genes in PMM-R parasite, respectively, while gray lines show the genes that are equally expressed in both samples and white regions represent those not hybridized to the probe. Chromosomes showing high up regulation in PMM resistance included Chromosome 12 and Chromosom 36 whereas high down regulation is evident on Chromosome 23. B. Distribution of genes differentially modulated during PMM resistance in Leishmania donovani according to gene ontology (GO) functional categories. Overall distribution of genes shows total number of unique genes up-regulated or down-regulated is 2.9%. GO categories of genes differentially expressed in PMM-R parasite suggest that genes belonging to metabolic process, electron transport chain/oxidation-reduction, cell surface proteins, stress proteins, cell movement and transporter activity were affected. Unclassified proteins include hypothetical proteins (proteins with unknown function and not tested experimentally) and proteins with no GO categories (unclassified) that have been experimentally characterized. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)