Figure 4. Loss of STRAP leads to epigenetic alterations in the promoter regions of NOTCH genes, and STRAP impairs the association between EZH2 and SUZ12 by interacting with SUZ12.
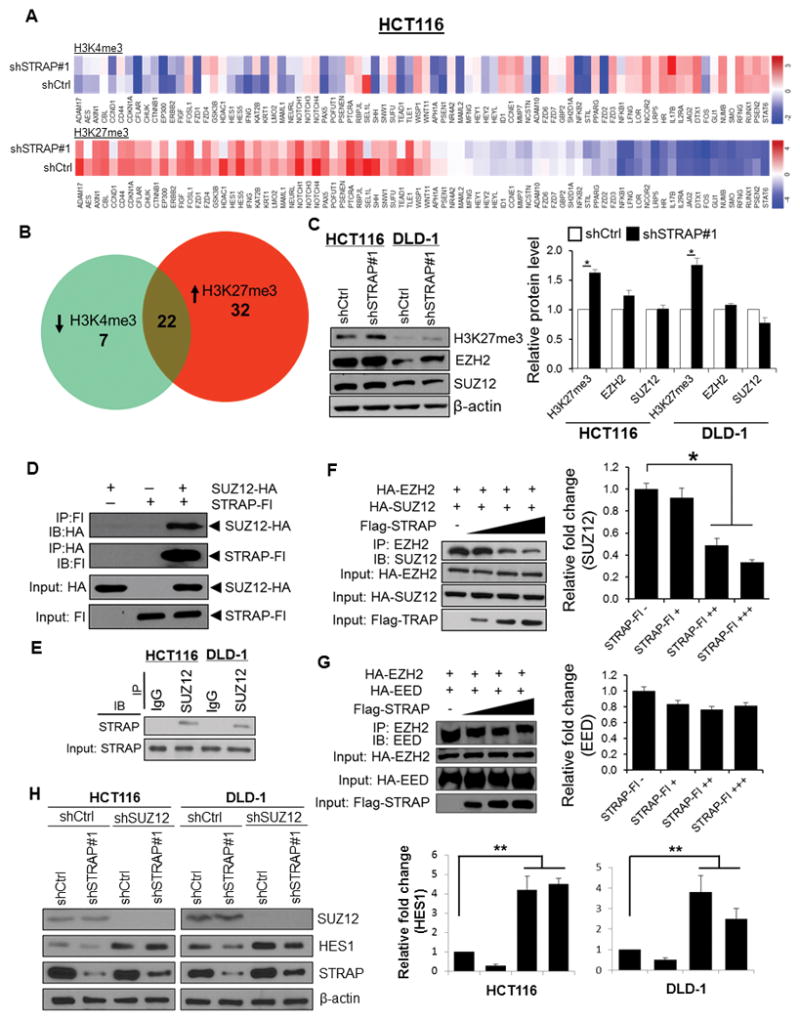
A. ChIP panel assays were performed to identify ChIP-enriched genomic DNA using antibodies against active chromatin marker H3K4me3 and repressive marker H3K27me3. Specifically designed qPCR primers focused on 1 kb genomic regions of individual promoter of 86 NOTCH signaling genes were used. Color bars in the heat maps indicate the altered fold from detected targets in shCtrl versus shSTRAP#1 HCT116 cells. B. Venn diagrams show the overlap of 22 genes with both low abundance of H3K4me3 (29 genes) and high abundance of H3K27me3 (54 genes) presented in KD cells relative to shCtrl parallel. C. Total cell lysates were analyzed for H3K27me3, EZH2, SUZ12, and β-actin expression by immunoblotting with their respective antibodies (left panel). Relative expressions of H3K27me3, EZH2 and SUZ12 normalized to β-actin were determined by densitometry using ImageJ software (right panel), *P<0.05, shSTRAP#1 vs. shCtrl. D. 293T cells were co-transfected with HA-tagged SUZ12 and Flag-tagged STRAP as indicated. Lysates were subjected to reciprocal immunoprecipitation with either anti-HA or anti-Flag antibody and analyzed by western blotting. Protein expression was also tested by immunoblotting. E. SUZ12 was immunoprecipitated with anti-SUZ12 antibody from lysates of indicated cell lines. Immune complexes were then analyzed by immunoblotting with anti-STRAP antibody. F and G. 293T cells were co-transfected with HA-tagged constructs of either SUZ12 or EED together with EZH2 as well as increased doses of Flag-tagged STRAP plasmid. Cell lysates were used for immunoprecipitation with the anti-EZH2 antibody. Co-precipitated SUZ12 (F) or EED (G) was detected by immunoblotting with the corresponding antibody (left panel). Protein expression was assessed by western blot. Relative fold changes of bound to SUZ12 (F) or EED (G) with increasing amounts of STRAP plasmid was determined by densitometry using ImageJ software (right panel). *P<0.05, when compared with no STRAP control. H. The indicated cells (shCtrl or shSTRAP#1) were infected with lentiviral shSUZ12RNA and scrambled shRNA. Lysates were subjected to western blotting with anti-SUZ12, anti-HES1, and anti-STRAP antibodies (left panel). β-actin was used as a loading control. Relative fold change of HES1 in each cell line was determined by densitometry using ImageJ software (right panel). **P<0.001, when compared with corresponding control.
