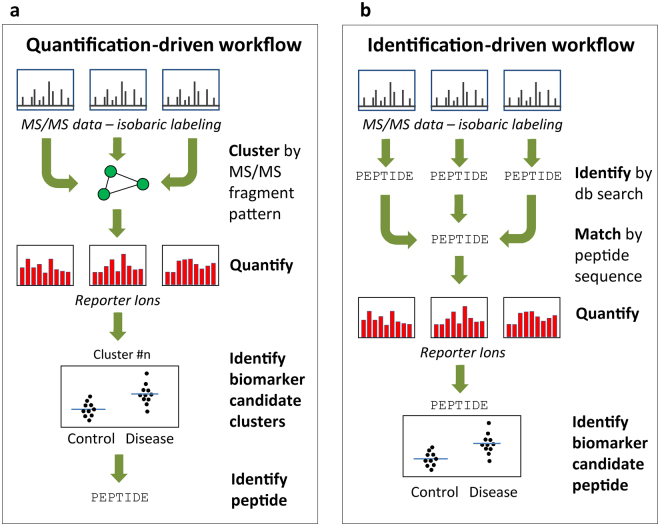
Figure 1.
Quantification-driven versus identification-driven proteomics. In the quantification-driven workflow (a), all MS/MS data, including data from different TMT sets and technical replicates, are clustered using an algorithm that matches spectra based on precursor ion m/z, charge state, and similarity of fragment ion pattern. For each cluster, the relative peptide abundance in the study participants, determined by the TMT reporter ion data, is used to single out biomarker candidate clusters, which are subsequently identified in targeted follow-up experiments. In contrast, in the identification-driven workflow (b), peptide identification is performed as the first step of data processing and the identified peptide sequences are used to match spectra for quantification. Consequently, only spectra that yield a positive identification can be included for biomarker evaluation.