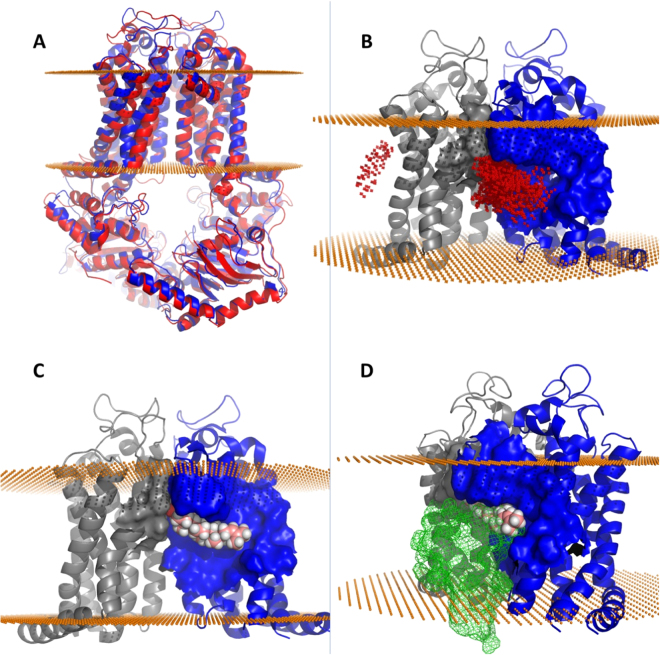
Figure 6.
Computed structural and binding features of the Abcg4 dimer model. Mouse Abcg4 sequence was submitted to the ModBase modeling server which constructed an Abcg4 model (blue) after selecting the x-ray crystal structure of ABCG5/ABCG8 (red cartoon) as the template in an unbiased search (Panel A). Panel B shows docking of desmosterol across the entire transmembrane surface identified two putative desmosterol binding sites (red dots represent the placement of desmosterol after each docking run). The overwhelming majority of final binding modes for desmosterol on Abcg4 were located in the homologous sterol binding cleft found in ABCG5/ABCG8 suggesting conserved sterol binding domain is found in both dimers. Best predicted binding mode of desmosterol (pink and white spheres) in the cleft formed by Abcg4 dimers (gray and blue, Panel C). Gray monomer rotated 90° away from the viewer along the axis of symmetry reveals competitive binding between desmosterol and Aβ (green mesh, panel D).