Abstract
Objective
Characterization of altered expression of selected transcripts linked to inflammation in the peripheral blood of sporadic amyotrophic lateral sclerosis (sALS) patients at early stage of disease to increase knowledge about peripheral inflammatory response in sALS.
Methods
RNA expression levels of 45 genes were assessed by RT-qPCR in 22 sALS cases in parallel with 13 age-matched controls. Clinical and serum parameters were assessed at the same time.
Results
Upregulation of genes coding for factors involved in leukocyte extravasation (ITGB2, INPP5D, SELL, and ICAM1) and extracellular matrix remodeling (MMP9 and TIMP2), as well as downregulation of certain chemokines (CCL5 and CXC5R), anti-inflammatory cytokines (IL10, TGFB2, and IL10RA), pro-inflammatory cytokines (IL-6), and T-cell regulators (CD2 and TRBC1) was found in sALS cases independently of gender, clinical symptoms at onset (spinal, respiratory, or bulbar), progression, peripheral leukocyte number, and integrity of RNA. MMP9 levels positively correlated with age, whereas CCR5, CCL5, and TRBC1 negatively correlated with age in sALS but not in controls. Relatively higher TNFA expression levels correlate with higher creatinine kinase protein levels in plasma.
Conclusion
Present findings show early inflammatory responses characterized by upregulation of factors enabling extravasation of leukocytes and extracellular matrix remodeling in blood in sALS cases, in addition to increased TNFA levels paralleling skeletal muscle damage.
Keywords: amyotrophic lateral sclerosis, blood, cytokines, extracellular matrix, leukocyte extravasation
Introduction
Increase in the number of astrocytes and microglia, and activation of inflammatory responses are major pathological marks in the anterior horn of the spinal cord in amyotrophic lateral sclerosis (ALS). Chronic inflammation plays the principal role in motor neuron demise and parallels the severity of motor neuron damage. A plethora of receptors, modulatory factors, chemokines, and anti- and pro-inflammatory cytokines are involved in this process at advanced stages of the disease (1–7). Inflammatory responses in the central nervous system are accompanied by modifications in blood and serum which may indicate a systemic inflammatory response in ALS (8–11). Peripheral nerves, autonomic nervous system, and muscle are involved in ALS, and they are putative targets of inflammatory reactions (12–18). Recent studies have also shown modifications in the intestinal microbiota in ALS (19, 20), thus categorizing ALS as a disease with multisystem involvement.
The majority of studies of blood and serum in ALS are at middle or advanced stages of the disease with or without treatment (10, 17, 21–26), but information about early stages at the time when the patient first asks for medical counseling and the disease is then diagnosed is limited (27). The purpose of the present study was to increase knowledge about expression of transcripts linked to inflammation in whole blood samples of sporadic ALS (sALS) patients at initial clinical stages of the disease. The selection of genes was conducted including representative pro- and anti-inflammatory cytokines, chemokines, cytokine modulators, extracellular matrix remodeling-related factors, molecules involved in extravasation mechanisms, oxidative stress markers, and T-cell regulators. The expression of these molecules was assessed considering the variables RNA integrity, gender, clinical symptoms at onset (spinal, respiratory, or bulbar), disease progression, peripheral leukocyte number, and creatinine kinase protein levels in plasma.
Materials and Methods
Sample Description
Whole peripheral blood samples for mRNA expression and biochemical studies were obtained within the two first months after the diagnosis. Samples were obtained from 22 sALS patients (mean age at plasma sampling 62.5 years; 16 men and 6 women) and 13 healthy age-matched controls (mean age at plasma sampling 65 years; 11 men and 4 women). sALS patients were selected on the basis of early stage at the diagnosis with homogenous parameters of gender, age, and treatment, whereas controls were recruited on the basis of homogenous parameters of gender and age. Patients were evaluated clinically according to the main signs at onset (spinal, bulbar, and respiratory) and categorized according to disease progression as fast, expected, and slow progression depending on the survival or the clinical evolution in those still alive. Fast progression was considered in patients who survived less than 3 years; expected progression was considered between 3 and 5 years, and slow for those still alive after 5 years. The ALS Functional Rating Scale Revised (ALS-FRS-R, version May 2015) was currently used in every case. No ALS cases or controls suffered from infection or inflammatory disorder at the time of sampling. None of them complained of systemic disease and none received any treatment related to ALS. No familial forms of ALS for C9ORF72, SOD1, TARDBP, and FUS mutations were detected when DNA of each patient was sequenced. Blood samples from sALS cases and age-matched controls were obtained following signed informed consent and approval by Clinical Research Ethics Committee (CEIC) of the Bellvitge University Hospital. A summary of cases is shown in Table 1.
Table 1.
Summary of cases analyzed in the present study.
| Case | Age at plasma sampling | Gender | Diagnosis | Initial symptoms | RIN value |
|---|---|---|---|---|---|
| 1 | 60 | M | Control | – | 9.1 |
| 2 | 68 | M | Control | – | 9.2 |
| 3 | 66 | F | Control | – | 9.0 |
| 4 | N/A | M | Control | – | 8.9 |
| 5 | 74 | M | Control | – | 8.0 |
| 6 | N/A | F | Control | – | 8.3 |
| 7 | 76 | F | Control | – | 7.8 |
| 8 | 67 | M | Control | – | 6.1 |
| 9 | 72 | F | Control | – | 6.0 |
| 10 | 44 | F | Control | – | 6.0 |
| 11 | 66 | F | Control | – | 6.1 |
| 12 | 62 | F | Control | – | 6.5 |
| 13 | 63 | F | Control | – | 6.0 |
| 14 | 60 | M | ALS | Spinal | 7.4 |
| 15 | 63 | M | ALS | Spinal | 8.7 |
| 16 | 66 | F | ALS | Bulbar | 8.9 |
| 17 | 53 | F | ALS | Bulbar | 7.3 |
| 18 | 73 | M | ALS | Bulbar | 8.6 |
| 19 | 65 | M | ALS | Spinal | 8.9 |
| 20 | 43 | M | ALS | Bulbar | 8.6 |
| 21 | 57 | F | ALS | Bulbar | 7.4 |
| 22 | 65 | M | ALS | Bulbar | 7.1 |
| 23 | 67 | M | ALS | Bulbar | 7.4 |
| 24 | 73 | M | ALS | Spinal | 6.1 |
| 25 | 73 | F | ALS | Spinal | 6.0 |
| 26 | 59 | F | ALS | Spinal | 8.7 |
| 27 | 65 | M | ALS | Respiratory | 7.1 |
| 28 | 42 | M | ALS | Bulbar | 9.2 |
| 29 | 75 | M | ALS | Respiratory | 8.1 |
| 30 | 75 | M | ALS | Bulbar | 7.9 |
| 31 | 29 | M | ALS | Spinal | 8.3 |
| 32 | 77 | M | ALS | Spinal | 7.4 |
| 33 | 55 | M | ALS | Spinal | 8.5 |
| 34 | 69 | M | ALS | Spinal | 8.6 |
| 35 | 71 | F | ALS | Spinal | 8.7 |
ALS, amyotrophic lateral sclerosis; M, male; F, female; RIN, RNA integrity number.
Blood Collection
In addition to current blood samples for hemogram and biochemical parameters, whole blood samples were collected using PAXgene Blood RNA Tube (PAXgene Blood RNA Tube, PreAnalytiX, Qiagen® GmbH, Hilden, GE) collecting system. Two PAXgene Blood RNA tubes were obtained per case. Samples were collected at the first visit once the clinical diagnosis was established. Tubes were kept for 2 h at room temperature to ensure lysis of blood cells and then stored at −20°C for 24 h. Thereafter, tubes were transferred to −80°C for at least 7 days prior to processing.
White Blood Cells (WBC) Counting
Blood was collected in EDTA 3 mL tubes and analyzed using flow-cytometry equipment. Technicon H-1, H-2, and H-3 apparatuses are discrete analyzers that perform complete blood and platelet counts, and leukocyte differential count. The instrument has a tungsten halogen light source and cytometer for leukocyte peroxidase analysis, with the addition of a helium-neon red laser for RBC/platelet and basophil determinations. Red blood cells are lysed, and fixed leukocytes flow in a stream sheath—a layer of inert liquid of the same refractive index. The stream sheath serves to narrow the sample stream, which prevents clogging and keeps the flow cell clean. Within the cell flow, cells are classified one by one on the basis of size (determined by a dark-field light scatter detector) and cytochemical peroxidase reaction. Measurement of the peroxidase activity is sufficient for most of the WBC differential classification. Lymphocytes are identified as small, unstained cells. Large atypical lymphocytes, plasma cells, and some blasts are characterized as “large unstained cells” (LUCs). Eosinophils exhibit the strongest peroxidase activity and appear smaller than neutrophils because they absorb some of their own scatter signal. Neutrophils are large and have moderate peroxidase activity. Monocytes have somewhat weaker peroxidase staining and are, therefore, in the area to the left of the neutrophils and to the right of the LUCs. The instrument’s computer automatically performs cluster analysis of the WBC subpopulations. The Technicon systems provide both relative (per cent) and absolute (×109 cells/L) cell counts for neutrophils, eosinophils, basophils, monocytes, and LUCs.
Quantitative Determination of Creatine Kinase (CK) in Blood Samples
Kinetic determination of CK was based upon IFCC (International Federation of Clinical Chemistry and Laboratory Medicine) and DGKC (Deutsche Gesellschaft für Klinische Chemie). The principle of the method is based on the ability of CK to catalyze the conversion of creatine phosphate and ADP to creatine and ATP. ATP and glucose are converted to ADP and glucose-6-phosphate by hexokinase. Glucose-6-phosphate dehydrogenase oxidizes glucose-6-phosphate to 6-phosphogluconate, reducing NADP to NADPH. The rate of conversion of NADP/NADPH, monitored at 340 nm, is proportional to CK activity. N-acetyl cysteine (NAC) is added as an activator of CK (28, 29).
RNA Extraction and RT-qPCR
PAXgene Blood RNA tubes were incubated overnight at 4°C in a shaker-plate to equilibrate the temperature and increase yields and then at room temperature for 2 h before starting the procedure. RNA from frozen whole blood samples was extracted following the instructions of the supplier (PAXgene Blood RNA kit, PreAnalytiX, Qiagen® GmbH, Hilden, GE). RNA integrity number (RIN) and 28S/18S ratios were determined with the Agilent Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA) to assess RNA quality. RNA concentration was evaluated using a NanoDrop™ Spectrophotometer (Thermo Fisher Scientific, Carlsbad, CA, USA). RIN values are shown in Table 1. Complementary DNA (cDNA) was prepared using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) following the protocol provided by the supplier. Parallel reactions for each RNA sample were run in the absence of MultiScribe Reverse Transcriptase to assess lack of genomic DNA contamination. TaqMan RT-qPCR assays were performed in duplicate for each gene on cDNA samples in 384-well optical plates using an ABI Prism 7900 Sequence Detection system (Applied Biosystems, Life Technologies, Waltham, MA, USA). For each 10 µL TaqMan reaction, 4.5 µL cDNA was mixed with 0.5 µL 20× TaqMan Gene Expression Assays and 5 µL of 2× TaqMan Universal PCR Master Mix (Applied Biosystems). The identification numbers and names of TaqMan probes are shown in Table 2. Probes were selected on the basis of our previous observations of inflammatory changes in the spinal cord and frontal cortex in sALS (7) together with additional markers linked to extravasation mechanisms and extracellular matrix remodeling. Mean values of two house-keeping genes, glucuronidase beta (GUS-β) (30) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (31), were used as internal controls for normalization. The reactions were carried out using the following parameters: 50°C for 2 min, 95°C for 10 min, and 40 cycles at 95°C for 15 s, and at 60°C for 1 min. Finally, all TaqMan PCR data were captured using the Sequence Detection Software (SDS version 2.2.2, Applied Biosystems). Samples were analyzed with the double-delta cycle threshold (ΔΔCT) method.
Table 2.
Genes, gene symbols, and references in the present series.
| Gene | Gene symbol | Reference |
|---|---|---|
| Catalase | CAT | Hs00156308_m1 |
| Cathepsin C | CTSC | Hs00175188_m1 |
| Cathepsin S | CTSS | Hs00356423_m1 |
| CD4 molecule/T-cell surface glycoprotein CD4 | CD4 | Hs01058407_m1 |
| CD44 molecule | CD44 | Hs01075861_m1 |
| CD8a molecule/T-cell surface glycoprotein CD8a Chain | CD8A | Hs00233520_m1 |
| Chemokine (C–C motif) ligand 5 | CCL5 | Hs00982282_m1 |
| Chemokine (C–C motif) receptor 5 | CCR5 | Hs00152917_m1 |
| Chemokine (C–X–C motif) receptor 5 | CXCR5 | Hs00173527_m1 |
| Colony stimulating factor 1 receptor | CSF1R | Hs00911250_m1 |
| Colony stimulating factor 3 receptor (granulocyte) | CSF3R | Hs00167918_m1 |
| C-type lectin domain family 7 member A | CLEC7A | Hs01124746_m1 |
| C–X–C motif chemokine ligand 8 | CXC8 | Hs00174103_m1 |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | Hs02786624_g1 |
| Inositol polyphosphate-5-phosphatase D | INPP5D | Hs00183290_m1 |
| Integrin subunit beta 2 | ITGB2 | Hs00164957_m1 |
| Integrin subunit beta 4 | ITGB4 | Hs00173995_m1 |
| Intercellular adhesion molecule 1 | ICAM-1 | Hs00164932_m1 |
| Intercellular adhesion molecule 5 | ICAM-5 | Hs00170285_m1 |
| Interferon, gamma | INFG | Hs00989291_m1 |
| Interleukin 1 beta | IL1B | Hs01555410_m1 |
| Interleukin 10 | IL10 | Hs00961622_m1 |
| Interleukin 10 receptor subunit alpha | IL10RA | Hs00155485_m1 |
| Interleukin 10 receptor subunit beta | IL10RB | Hs00988697_m1 |
| Interleukin 6 | IL6 | Hs00985639_m1 |
| Interleukin 6 signal transducer | IL6ST | Hs00174360_m1 |
| LFA-3 receptor | CD2 | Hs00233515_m1 |
| Lymphocyte function-associated antigen 1 | LFA-1 | Hs00158218_m1 |
| Macrophage inflammatory protein 1-alpha | CCL3 | Hs00234142_m1 |
| Membrane-associated ring finger (C3HC4) 9 | MARCH9 | Hs04189729_m1 |
| Monocyte chemotactic and activating factor | CCL2 | Hs00234140_m1 |
| Metallopeptidase-9 | MMP9 | Hs00234579_m1 |
| Osteopontin | SPP1 | Hs00959010_m1 |
| Programmed cell death 1 ligand 2 | PD1L2 | Hs01057777_m1 |
| Selectin L | SELL | Hs00174151_m1 |
| Superoxide dismutase 1, soluble | SOD1 | Hs00533490_m1 |
| Superoxide dismutase 2, mitochondrial | SOD2 | Hs00167309_m1 |
| T cell receptor beta constant 1 | TRBC1 | Hs01588269_g1 |
| TIMP metallopeptidase inhibitor 1 | TIMP-1 | Hs00171558_m1 |
| TIMP metallopeptidase inhibitor 2 | TIMP-2 | Hs01091317_m1 |
| Toll-like receptor 2 | TLR2 | Hs00610101_m1 |
| Toll-like receptor 3 | TLR3 | Hs01551078_m1 |
| Toll-like receptor 4 | TLR4 | Hs01060206_m1 |
| Toll-like receptor 7 | TLR7 | Hs00152971_m1 |
| Tumor growth factor B1 | TGFB1 | Hs00998133_m1 |
| Tumor growth factor B2 | TGFB2 | Hs00234244_m1 |
| Tumor necrosis factor receptor superfamily member 1A | TNFRSF1 | Hs01042313_m1 |
| Tumor necrosis factor-alpha | TNFA | Hs01113624_g1 |
| Vascular endothelial growth factor A | VEGFA | Hs00900055_m1 |
| β-Glucuronidase | GUS-β | Hs00939627_m1 |
Statistical Analysis
The normality of distribution of fold change values was analyzed with the Kolmogorov–Smirnov test. The non-parametric Mann–Whitney test was performed to compare each group when values did not follow a normal distribution, whereas the unpaired t-test was used for normal variables. Statistical analysis and graphic design were performed with GraphPad Prism version 5.01 (La Jolla, CA, USA). Results were analyzed with Student’s t-test. Outliers were detected using the GraphPad software QuickCalcs (p < 0.05). The data were expressed as mean ± SEM and significance levels were set at *p < 0.05 and **p < 0.01 and ***p < 0.001, and tendencies at #<0.1. Pearson’s correlation coefficient was used to assess a possible linear association between two continuous quantitative variables.
Results
General Clinical and Hematological Findings
Amyotrophic lateral sclerosis progression was heterogeneous in the present series. Hemogram was not altered in sALS patients with the exception of a few cases in whom slight increase of neutrophils and low levels of lymphocytes was observed. CK levels were out of range in some patients and moderately increased in a few sALS cases. Clinical, hematological, and biochemical data are summarized in Table 3.
Table 3.
Biochemical alterations in blood samples of sporadic amyotrophic lateral sclerosis (sALS) cases.
| sALS case | Clinical progression | Creatinine kinase (CK) (μkat/L) | Leukocyte populations (×10E9cells/L) |
||||
|---|---|---|---|---|---|---|---|
| Neutrophil (1.5–5.7) | Lymphocyte (1.3–3.4) | Monocyte (0.31–0.92) | Eosinophil (0.03–0.39) | Basophil (0.01–0.09) | |||
| 14 | Expected | 13.9a (≤4.50) | 3.7 | 1.4 | 0.48 | 0.02b | 0.02 |
| 15 | Expected | 5.5a (≤4.50) | 3.4 | 2.1 | 0.46 | 0.21 | 0.04 |
| 16 | Expected | 3.5a (≤2.30) | 6.9a | 0.8b | 0.37 | 0.04 | 0.04 |
| 17 | Fast | 5.0a (≤2.30) | 4.2 | 1.0b | 0.34 | 0.1 | 0.05 |
| 18 | Fast | 3.5 (≤4.50) | N/A | N/A | N/A | N/A | N/A |
| 19 | Slow | 0.8 (≤4.50) | 4.3 | 1.2 | 0.44 | 0.16 | 0.04 |
| 20 | Fast | 4.6a (≤4.50) | N/A | N/A | N/A | N/A | N/A |
| 21 | Expected | 5.9a (≤4.50) | 3.3 | 1.0b | 0.30b | 0.15 | 0.04 |
| 22 | Expected | 2.2 (≤4.50) | 3.9 | 2.5 | 0.56 | 0.34 | 0.01 |
| 23 | Expected | 2.6 (≤4.50) | 7.8a | 1.0b | 0.6 | 0.01b | 0.03 |
| 24 | Expected | 2.8 (≤4.50) | 6.6a | 1.8 | 0.6 | 0.15 | 0.06 |
| 25 | Fast | 0.7 (≤4.50) | 5.6 | 1.4 | 0.87 | 0.39 | 0.09 |
| 26 | Fast | N/A | 3.6 | 1.6 | 0.49 | 0.11 | 0.02 |
| 27 | Expected | 8.3a (≤4.50) | 7.0a | 1.7 | 0.53 | 0.11 | 0.07 |
| 28 | Fast | 1.8 (≤4.50) | 4.2 | 3.1 | 0.76 | 0.28 | 0.04 |
| 29 | Fast | N/A | 6.0a | 0.9b | 0.74 | 0.08 | 0.03 |
| 30 | Fast | 3.0 (≤4.50) | 4.3 | 1.4 | 0.77 | 0.04 | 0.04 |
| 31 | Fast | 2.1 (≤4.50) | 4.2 | 2.1 | 0.63 | 0.19 | 0.08 |
| 32 | Fast | N/A | N/A | N/A | N/A | N/A | N/A |
| 33 | Slow | 2.0 (≤4.50) | 3.8 | 2.5 | 0.44 | 0.55 | 0.06 |
| 34 | Slow | 3.5 (≤4.50) | 3.5 | 1.8 | 0.38 | 0.19 | 0.06 |
| 35 | Fast | 11.6a (≤4.50) | N/A | N/A | N/A | N/A | N/A |
N/A, data not available; μkat/L, microkatals/liter.
Normal CK levels in brackets (these are variable depending on the method used; CK values in every ALS case are evaluated according to the method used).
aAbove normal range.
bBelow normal range.
Gene Expression Levels
Anti-inflammatory Cytokines
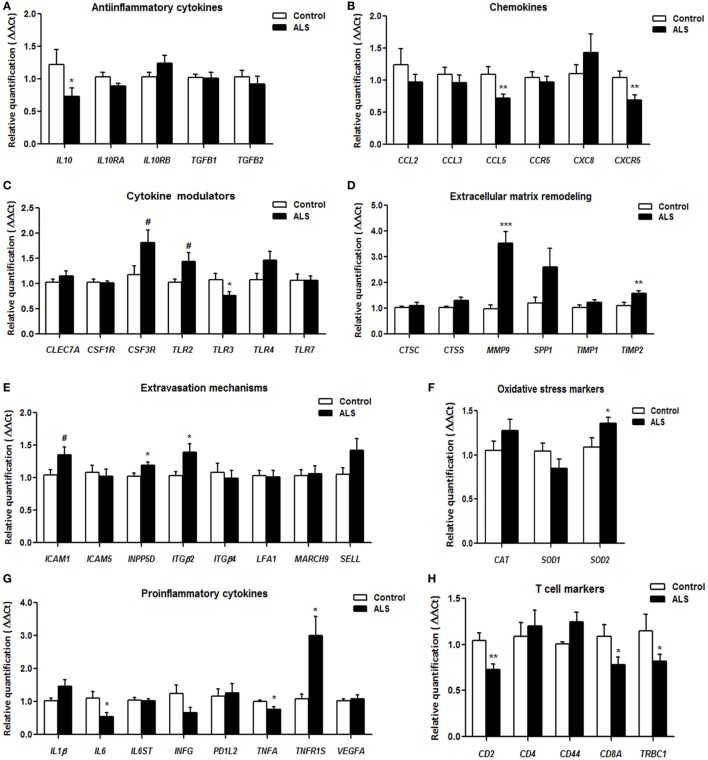
IL10, coding for interleukin 10, and TGFB2, coding for transforming growth factor beta 1, mRNA levels were significantly reduced in sALS, whereas IL10RA which codes for interleukin 10 receptor subunit alpha showed a tendency to decrease. Expression levels of IL10RB and TGFB1 encoding interleukin 10 receptor subunit beta and transforming growth factor beta 1, respectively, were not modified (Figure 1A).
Figure 1.
Gene expression of anti-inflammatory cytokines (A), chemokines (B), cytokine modulators (C), extracellular matrix remodeling-related factors (D), molecules involved in extravasation mechanisms (E), oxidative stress markers (F), pro-inflammatory cytokines (G), and T-cell markers (H), as revealed by RT-qPCR, in blood from control and sporadic amyotrophic lateral sclerosis (sALS) cases. All data were expressed as the mean ± SEM. Statistical comparisons were performed using unpaired t-test; significance level was set at *p < 0.05, **p < 0.01 and ***p < 0.001, and tendencies at #<0.1. A total of 13 healthy samples and 22 sALS samples were included in RT-qPCR analysis.
Chemokines
Expression levels of CCL5 and CXC5R, which code for C-C motif chemokine ligand 5 and C-X-C motif chemokine receptor 5, respectively, were significantly decreased; CCR5 coding for C-C motif chemokine receptor 5 showed a tendency to decrease. No modifications were seen for C-C motif chemokine ligand 2 (CCL2) and 3 (CCL3), and C-X-C motif chemokine 8 (CXC8) (Figure 1B).
Cytokine Modulators
Toll like receptors TLR2 and TLR4 mRNA expression showed a tendency to increase, whereas TLR3 mRNA expression was significantly decreased in sALS. TLR7 and other genes involved in cytokine modulation such as C-type lectin domain family 7 member A (CLEC7A), colony stimulating factor 1 receptor (CSF1R), and colony stimulating factor 3 receptor (CSF3R) were not altered (Figure 1C).
Extracellular Matrix Remodeling
MMP9, coding for matrix metallopeptidase 9, and TIMP2, coding for its inhibitor protein, TIMP metallopeptidase inhibitor 2, were significantly increased in sALS. The expression levels of CTSC, CTSS, TIMP1, and SPP1, coding for cathepsin C, cathepsin S, TIMP metallopeptidase inhibitor 1, and osteopontin, respectively, were similar in sALS and controls (Figure 1D).
Extravasation Mechanisms
ITGB2, coding for integrin subunit beta 2, and INPP5D, coding for inositol polyphosphate-5-phosphatase D, were upregulated in sALS. Tendency to increase was found for SELL and ICAM1, coding for selectin-L and intercellular adhesion molecule 1, respectively. No changes were detected in the expression of ICAM5, ITGB4, LFA1, and MARCH9 encoding, respectively, intercellular adhesion molecule 5, integrin subunit beta 4, lymphocyte function-associated antigen 1, and membrane associated ring-CH-type finger 9 (Figure 1E).
Oxidative Stress Markers
Expression of catalase (CAT) and superoxide dismutase 1 (SOD1) genes was not modified. Superoxide dismutase 2 (SOD2) showed a tendency to increase in sALS (Figure 1F).
Pro-inflammatory Cytokines
IL6, coding for interleukin-6, was significantly downregulated in sALS cases. TNF-α coding gene TNFA showed a tendency to decrease. In contrast, TNFR1S, the gene coding for its receptor, was significantly increased. No alterations were found in the remaining assessed genes IL1B, IL6ST, INFG, PD1L2, and VEGFA, coding for interleukin 1B, interleukin 6 signal transducer, interferon gamma, programmed cell death 1 ligand 2, and vascular endothelial growth factor A, respectively (Figure 1G).
T Cell Markers
Expression of CD2, coding for CD2 molecule; CD8A, coding for T-Cell Surface Glycoprotein CD8 Alpha Chain; and TRBC1, coding for T-cell receptor beta constant 1, was significantly decreased in sALS cases. The expression of CD44 and T-cell surface glycoprotein CD4 gene (CD4) was not modified (Figure 1H).
Correlation between Clinical Parameters and Gene Transcription
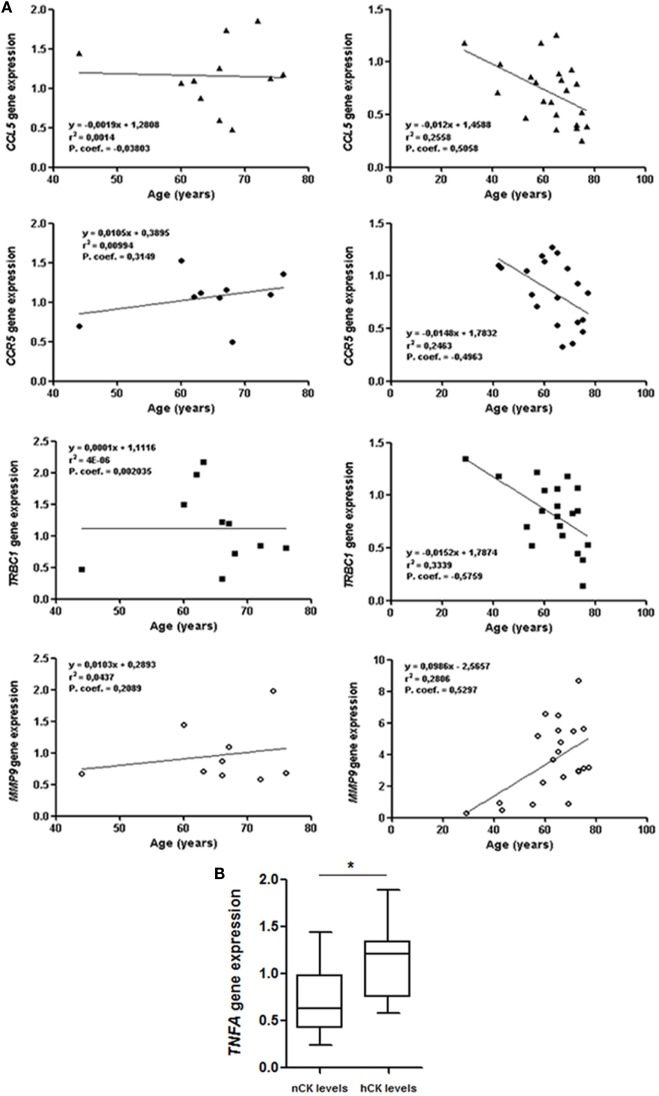
Gender, ALS form of onset (spinal, bulbar, and respiratory), clinical progression, leukocyte counts and leukocyte types, and RIN values did not correlate with modifications in gene expression. However, MMP9 levels in sALS cases positively correlated with age (p = 0.046) (Figure 2A). CCR5 (p = 0.0307), CCL5 (p = 0.016), and TRBC1 (p = 0.0076) negatively correlated with age (Figure 2A) in sALS. These changes were not observed in the control group. Importantly, patients with sALS showed significant relation between elevated levels of TNFA gene and creatinine kinase (CK) values, which were out of the normal range (p = 0.025) (Figure 2B).
Figure 2.
(A) Positive correlation between MMP9 and age at sampling, and negative correlation between age at sampling and CCL5, CCR5, and TRBC1 in sporadic amyotrophic lateral sclerosis (sALS) (right graphs), but not in control cases (left graphs). (B) Relation between TNFA mRNA expression levels in blood and creatine kinase (CK) protein levels in serum in sALS (nCK, normal CK levels; hCK, high/out of range CK levels) using Student’s t-test. Gene expression values correspond to fold change values of ΔΔCT.
Discussion
Peripheral inflammatory responses are common, but poorly defined, in human neurodegenerative diseases. Several studies focus on inflammatory responses in spinal cord and blood in sALS (1–11). The present study was geared to gain information about inflammatory gene expression profiles in the whole blood in a series of sALS patients at the beginning of clinical symptoms and non-treated with riluzole in order to avoid bias related to the treatment.
Present observations complement data from previous studies and point to the activation of mechanisms facilitating extravasation of WBC to target organs.
Neutrophil recruitment is supported by leukocyte adhesion molecules, chemokines, and cytokines (32, 33). Increased expression of ITGB2 and a tendency of ICAM1 to increase in blood suggest that adhesion and trans-endothelial migration of leukocytes is facilitated in sALS (34–36). Selectin 1, encoded by SELL, participates in leukocyte binding to endothelial cells and facilitates migration of WBC (37, 38); SELL expression has a tendency to increase in sALS. Increased expression of MMP9 favors degradation of extracellular matrix components and facilitation of leukocyte migration (39). MMP9 is usually secreted in conjunction with TIMP-1, a specific inhibitor, which controls its proteolytic activity (40). A balance between MMP9 and TIMP-1 proteins regulates excessive tissue degradation in chronic inflammation (41). However, mRNA expression levels of cathepsins, also involved in extracellular matrix degradation (42), are not modified in blood of ALS cases when compared with blood samples from controls.
Expression levels of CCL2 and CCL3 v, the products of which modulate monocyte attraction (43, 44) are not modified in sALS. Moreover, reduced expression of CCR5, CCL5, and CXCR5 supports reduced activation of B-cells (45).
The product of CD2 expressed in T-cells modulates T-cell proliferation (46), whereas the product of TRBC1 is implicated in T-cell activation (47). CCL5 and CCR5 encode T-cell chemo-attractant and regulatory molecules (48, 49). Reduced mRNA expression of these markers suggests inhibition of T-cell signaling.
Finally, increased INPP5D mRNA expression favors a negative regulation of myeloid cell proliferation (50).
Toll-like receptors are involved in the initiation of the inflammatory process (51). Reduced levels of TLR3 accompanied by tendency to increased TLR4 and TLR2 mRNA expression point to ambiguous activation signaling by Toll-like receptors.
TGFB2, IL10, and IL6 mRNAs are downregulated, and IL10RA and TNFA have tendency to decrease in blood in sALS when compared with controls. Expression levels of IL10RB, TGFB1, IL1β, IL6ST, INFG (coding for interferon γ), and VEGFA are not modified in sALS. Expression levels of assessed colony-stimulating receptors and CSF3R do not differ from control values. Even considering the increased expression of TNFR1S mRNA, the final scenario is downregulation of pro- and anti-inflammatory cytokines in sALS.
SOD1 transgenic mice lacking functional CD4+ T cells show increased motor neuron damage which is reversed following bone marrow transplants thus suggesting a neuroprotective role of CD4+ T cells (52). On the other hand, SOD1 transgenic mice with additional depletion of the Rag2 gene (mSOD1/RAG2−/− mice) show delayed motor neuron disease, thus suggesting that mature lymphocytes produce deleterious effects on vulnerable motor neurons (53).
Previous studies have shown a higher percentage of IL-13-positive CD4 and CD8 lymphocytes (8), increased numbers of peripheral CD8 cytotoxic T-cells and natural killer cells, together with decreased regulatory T (treg) lymphocytes (10) in ALS. Our observations show decreased expression of CD2, coding for CD2 molecule, TRBC1, coding for T-cell receptor beta constant 1 and CD8 mRNA, and preserved CD4 mRNA expression. Therefore, additional studies are necessary to elucidate these discrepancies in larger series.
The present findings show a complex scenario at early clinical stages of sALS, including on the one hand upregulation of genes whose products are involved in leukocyte extravasation and extracellular matrix remodeling, and on the other, downregulation of chemokines, anti- and pro-inflammatory cytokines, and lymphocyte modulators.
Positive correlation between MMP9 and age, and negative correlation between age and CCL5, CCR5, and TRBC1 has been observed in sALS but not in controls. No correlation has been found between present observations and first clinical manifestation, gender, and disease progression. Therefore, the present findings have little prognosis value.
There is only positive correlation between TNFA mRNA expression and CK levels. Although TNFA mRNA expression is lower in ALS when compared with controls, higher TNFA mRNA values correlate with higher CK protein levels. This observation points to the possibility of a link between TNFA and muscular damage in sALS. Previous studies have shown that muscular pathology is accompanied by increased expression of systemic inflammatory markers (17). Moreover, increased expression of inflammatory markers, including IL-1β and TNF-α, is found in the skeletal muscle at symptomatic and end-stages of SOD1(G93A) transgenic mice (18). However, these individual data are not sufficient to advance any definitive conclusion.
Transcriptome studies at early clinical stages in SOD1(G93A) transgenic mice have shown deregulated pathways common to spinal cord, muscle and sciatic nerve; two pathways are associated with T cell activation, two with macrophage activation, and one pathway contains genes involved in co-stimulatory regulation of the adaptive and innate immune systems; but blood did not show representation of these altered pathways (54). However, genetic ablation of IP3 receptor 2, which modulates inflammation and which expression is augmented in the spinal cord in ALS and related mice models, increases cytokines and decreases survival of SOD1G93A mice (55). These studies point to involvement of peripheral blood cells in the inflammatory response in the spinal cord in ALS. Present observations show systemic inflammatory responses linked to extravasation of leukocytes and remodeling of extracellular matrix at early stages of sALS. However, the observed changes do not indicate the primary or secondary origin, and the precise link between intrinsic and peripheral inflammatory responses in the pathogenesis of sALS.
Ethics Statement
Blood samples from sALS cases and age-matched controls were obtained following signed informed consent and approval by Clinical Research Ethics Committee (CEIC) of the Bellvitge University Hospital.
Author Contributions
All the authors designed, supervised the study, and wrote the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank T. Yohannan for editorial help.
Footnotes
Funding. This study was supported by grants from CIBERNED and Instituto de Salud Carlos III, and co-funded by FEDER funds/European Regional Development Fund (ERDF)—a way to build Europe; ALS intra-CIBERNED project to IF and IFI15/00035 fellowship to PA-B.
References
- 1.Graves MC, Fiala M, Dinglasan LA, Liu NQ, Sayre J, Chiappelli F, et al. Inflammation in amyotrophic lateral sclerosis spinal cord and brain is mediated by activated macrophages, mast cells and T cells. Amyotroph Lateral Scler Other Motor Neuron Disord (2004) 5:213–9. 10.1080/14660820410020286 [DOI] [PubMed] [Google Scholar]
- 2.Henkel JS, Engelhardt JI, Siklós L, Simpson EP, Kim SH, Pan T, et al. Presence of dendritic cells, MCP-1, and activated microglia/macrophages in amyotrophic lateral sclerosis spinal cord tissue. Ann Neurol (2004) 55:221–35. 10.1002/ana.10805 [DOI] [PubMed] [Google Scholar]
- 3.Johann S, Heitzer M, Kanagaratnam M, Goswami A, Rizo T, Weis J, et al. NLRP3 inflammasome is expressed by astrocytes in the SOD1 mouse model of ALS and in human sporadic ALS patients. Glia (2015) 63:2260–73. 10.1002/glia.22891 [DOI] [PubMed] [Google Scholar]
- 4.Casula M, Iyer AM, Spliet WG, Anink JJ, Steentjes K, Sta M, et al. Toll-like receptor signaling in amyotrophic lateral sclerosis spinal cord tissue. Neuroscience (2011) 179:233–43. 10.1016/j.neuroscience.2011.02.001 [DOI] [PubMed] [Google Scholar]
- 5.Sta M, Sylva-Steenland RM, Casula M, de Jong JM, Troost D, Aronica E, et al. Innate and adaptive immunity in amyotrophic lateral sclerosis: evidence of complement activation. Neurobiol Dis (2011) 42:211–20. 10.1016/j.nbd.2011.01.002 [DOI] [PubMed] [Google Scholar]
- 6.Puentes F, Malaspina A, Van Noort JM, Amor S. Non-neuronal cells in ALS: role of glial, immune cells and blood-CNS barriers. Brain Pathol (2016) 26:248–57. 10.1111/bpa.12352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrés-Benito P, Moreno J, Aso E, Povedano M, Ferrer I. Amyotrophic lateral sclerosis, gene deregulation in the anterior horn of the spinal cord and frontal cortex area 8: implications in frontotemporal lobar degeneration. Aging (Albany NY) (2017) 9(3):823–51. 10.18632/aging.101195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi N, Kawano Y, Tateishi T, Kikuchi H, Osoegawa M, Ohyagi Y, et al. Increased IL-13-producing T cells in ALS: positive correlations with disease severity and progression rate. J Neuroimmunol (2007) 182:232–5. 10.1016/j.jneuroim.2006.10.001 [DOI] [PubMed] [Google Scholar]
- 9.Rentzos M, Evangelopoulos ME, Sereti E, Zouvelou V, Marmara S, Alexakis T, et al. Humoral immune activation in amyotrophic lateral sclerosis patients. Neurol Int (2013) 5:e3. 10.4081/ni.2013.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rentzos M, Evangelopoulos E, Sereti E, Zouvelou V, Marmara S, Alexakis T, et al. Alterations of T cell subsets in ALS: a systemic immune activation? Acta Neurol Scand (2012) 125:260–4. 10.1111/j.1600-0404.2011.01528.x [DOI] [PubMed] [Google Scholar]
- 11.Henkel JS, Beers DR, Wen S, Rivera AL, Toennis KM, Appel JE, et al. Regulatory T-lymphocytes mediate amyotrophic lateral sclerosis progression and survival. EMBO Mol Med (2013) 5:64–79. 10.1002/emmm.201201544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piccione EA, Sletten DM, Staff NP, Low PA. Autonomic system and amyotrophic lateral sclerosis. Muscle Nerve (2015) 51:676–9. 10.1002/mus.24457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian F, Yang W, Mordes DA, Wang JY, Salameh JS, Mok J, et al. Monitoring peripheral nerve degeneration in ALS by label-free stimulated Raman scattering imaging. Nat Commun (2016) 7:13283. 10.1038/ncomms13283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiber S, Dannhardt-Stieger V, Henkel D, Debska-Vielhaber G, Machts J, Abdulla S, et al. Quantifying disease progression in amyotrophic lateral sclerosis using peripheral nerve sonography. Muscle Nerve (2016) 54:391–7. 10.1002/mus.25066 [DOI] [PubMed] [Google Scholar]
- 15.Vucic S. Sensory and autonomic nervous system dysfunction in amyotrophic lateral sclerosis. Neuropathol Appl Neurobiol (2017) 43(2):99–101. 10.1111/nan.12336 [DOI] [PubMed] [Google Scholar]
- 16.Nolano M, Provitera V, Manganelli F, Iodice R, Caporaso G, Stancanelli A, et al. Non-motor involvement in amyotrophic lateral sclerosis: new insight from nerve and vessel analysis in skin biopsy. Neuropathol Appl Neurobiol (2016) 43(2):119–32. 10.1111/nan.12332 [DOI] [PubMed] [Google Scholar]
- 17.Lu CH, Allen K, Oei F, Leoni E, Kuhle J, Tree T, et al. Systemic inflammatory response and neuromuscular involvement in amyotrophic lateral sclerosis. Neurol Neuroimmunol Neuroinflamm (2016) 3:e244. 10.1212/NXI.0000000000000244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Dyke JM, Smit-Oistad IM, Macrander C, Krakora D, Meyer MG, Suzuki M. Macrophage-mediated inflammation and glial response in the skeletal muscle of a rat model of familial amyotrophic lateral sclerosis (ALS). Exp Neurol (2016) 277:275–82. 10.1016/j.expneurol.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang X. Potential role of gut microbiota and tissue barriers in Parkinson’s disease and amyotrophic lateral sclerosis. Int J Neurosci (2016) 126:771–6. 10.3109/00207454.2015.1096271 [DOI] [PubMed] [Google Scholar]
- 20.Zhang YG, Wu S, Yi J, Xia Y, Jin D, Zhou J, et al. Target intestinal microbiota to alleviate disease progression in amyotrophic lateral sclerosis. Clin Ther (2017) 39:322–36. 10.1016/j.clinthera.2016.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang R, Gascon R, Miller RG, Gelinas DF, Mass J, Lancero M, et al. MCP-1 chemokine receptor CCR2 is decreased on circulating monocytes in sporadic amyotrophic lateral sclerosis (sALS). J Neuroimmunol (2006) 179(1–2):87–93. 10.1016/j.jneuroim.2006.06.008 [DOI] [PubMed] [Google Scholar]
- 22.Mantovani S, Garbelli S, Pasini A, Alimonti D, Perotti C, Melazzini M, et al. Immune system alterations in sporadic amyotrophic lateral sclerosis patients suggest an ongoing neuroinflammatory process. J Neuroimmunol (2009) 210(1–2):73–9. 10.1016/j.jneuroim.2009.02.012 [DOI] [PubMed] [Google Scholar]
- 23.Zhao W, Beers DR, Hooten KG, Sieglaff DH, Zhang A, Kalyana-Sundaram S, et al. Characterization of gene expression phenotype in amyotrophic lateral sclerosis monocytes. JAMA Neurol (2017) 74(6):677–85. 10.1001/jamaneurol.2017.0357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeda J, Kohriyama T, Nakamura S. Elevation of serum soluble E-selectin and antisulfoglucuronyl paragloboside antibodies in amyotrophic lateral sclerosis. Eur J Neurol (2000) 7(5):541–7. 10.1046/j.1468-1331.2000.t01-1-00114.x [DOI] [PubMed] [Google Scholar]
- 25.Babu GN, Kumar A, Chandra R, Puri SK, Kalita J, Misra UK. Elevated inflammatory markers in a group of amyotrophic lateral sclerosis patients from northern India. Neurochem Res (2008) 33(6):1145–9. 10.1007/s11064-007-9564-x [DOI] [PubMed] [Google Scholar]
- 26.Cereda C, Baiocchi C, Bongioanni P, Cova E, Guareschi S, Metelli MR, et al. TNF and sTNFR1/2 plasma levels in ALS patients. J Neuroimmunol (2008) 194(1):123–31. 10.1016/j.jneuroim.2007.10.028 [DOI] [PubMed] [Google Scholar]
- 27.Waller R, Goodall EF, Milo M, Cooper-Knock J, Da Costa M, Hobson E, et al. Serum miRNAs miR-206, 143-3p and 374b-5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol Aging (2017) 55:123–31. 10.1016/j.neurobiolaging.2017.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hørder M, Elser RC, Gerhardt W, Mathieu M, Sampson EJ. Approved recommendation of IFCC methods for the measurement of catalytic concentration of enzymes, part 7 IFCC method for creatine kinase. Eur J Clin Chem Clin Biochem (1991) 29:435. [PubMed] [Google Scholar]
- 29.Young DS, Friedman RB. Effects of Disease on Clinical Laboratory Tests. 4th ed Washington, DC: AACC Press; (2001). [Google Scholar]
- 30.Zampieri M, Ciccarone F, Guastafierro T, Bacalini MG, Calabrese R, Moreno-Villanueva M, et al. Validation of suitable internal control genes for expression studies in aging. Mech Ageing Dev (2010) 131:89–95. 10.1016/j.mad.2009.12.005 [DOI] [PubMed] [Google Scholar]
- 31.Bayatti N, Cooper-Knock J, Bury JJ, Wyles M, Heath PR, Kirby J, et al. Comparison of blood RNA extraction methods used for gene expression profiling in amyotrophic lateral sclerosis. PLoS One (2014) 9:e87508. 10.1371/journal.pone.0087508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ley K. Integration of inflammatory signals by rolling neutrophils. Immunol Rev (2002) 186:8–18. 10.1034/j.1600-065X.2002.18602.x [DOI] [PubMed] [Google Scholar]
- 33.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol (2006) 6:173–82. 10.1038/nri1785 [DOI] [PubMed] [Google Scholar]
- 34.Smith CW, Marlin SD, Rothlein R, Toman C, Anderson DC. Cooperative interactions of LFA-1 and MAC-1 with intercellular adhesion molecule-1 in facilitating adherence and transendothelial migration of human neutrophils in vitro. J Clin Invest (1989) 83:2008–17. 10.1172/JCI114111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefort CT, Ley K. Neutrophil arrest by LFA-1 activation. Front Immunol (2012) 3:157. 10.3389/fimmu.2012.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hua S. Targeting sites of inflammation: intercellular adhesion molecule-1 as a target for novel inflammatory therapies. Front Pharmacol (2013) 4:127. 10.3389/fphar.2013.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol (2007) 7:678–89. 10.1038/nri2156 [DOI] [PubMed] [Google Scholar]
- 38.Marki A, Esko JD, Pries AR, Ley K. Role of the endothelial surface layer in neutrophil recruitment. J Leukoc Biol (2015) 98:503–15. 10.1189/jlb.3MR0115-011R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandooren J, Van den Steen PE, Opdenakker G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): the next decade. Crit Rev Biochem Mol Biol (2013) 48:222–72. 10.3109/10409238.2013.770819 [DOI] [PubMed] [Google Scholar]
- 40.Kurzepa J, Madro A, Czechowska G, Kurzepa J, Celiński K, Kazmierak W, et al. Role of MMP-2 and MMP-9 and their natural inhibitors in liver fibrosis, chronic pancreatitis and non-specific inflammatory bowel diseases. Hepatobiliary Pancreat Dis Int (2014) 13:570–9. 10.1016/S1499-3872(14)60261-7 [DOI] [PubMed] [Google Scholar]
- 41.Amalinei C, Caruntu ID, Giuşca SE, Balan RA. Matrix metalloproteinases involvement in pathologic conditions. Rom J Morphol Embryol (2010) 51:215–28. [PubMed] [Google Scholar]
- 42.Fonović M, Turk B. Cysteine cathepsins and extracellular matrix degradation. Biochim Biophys Acta (2014) 1840:2560–70. 10.1016/j.bbagen.2014.03.017 [DOI] [PubMed] [Google Scholar]
- 43.Paavola CD, Hemmerich S, Grunberger D, Polsky I, Bloom A, Freedman R, et al. Monomeric monocyte chemoattractant protein-1 (MCP-1) binds and activates the MCP-1 receptor CCR2B. J Biol Chem (1998) 273:33157–65. 10.1074/jbc.273.50.33157 [DOI] [PubMed] [Google Scholar]
- 44.Sanadgol N, Golab F, Mostafaie A, Mehdizadeh M, Abdollahi M, Sharifzadeh M, et al. Ellagic acid ameliorates cuprizone-induced acute CNS inflammation via restriction of microgliosis and down-regulation of CCL2 and CCL3 pro-inflammatory chemokines. Cell Mol Biol (2016) 62:24–30. 10.14715/cmb/2016.62.12.5 [DOI] [PubMed] [Google Scholar]
- 45.Le Y, Zhou Y, Iribarren P, Wang J. Chemokines and chemokine receptors: their manifold roles in homeostasis and disease. Cell Mol Immunol (2004) 1:95–104. [PubMed] [Google Scholar]
- 46.The SO, Killeen N, Tarakhovsky A, Littman DR, The HS. CD2 regulates the positive selection and function of antigen-specific CD4-CD8 T cells. Blood (1997) 89:1308–18. [PubMed] [Google Scholar]
- 47.MacLean SJ, Gibson DM. Identification of a predominant sequence variant of the T-cell receptor TCRBC1 gene. Immunogenetics (1997) 45:223–5. 10.1007/s002510050194 [DOI] [PubMed] [Google Scholar]
- 48.Siveke JT, Hamann A. T helper 1 and T helper 2 cells respond differentially to chemokines. J Immunol (1998) 160:550–4. [PubMed] [Google Scholar]
- 49.Rabin RL, Park MK, Liao F, Swofford R, Stephany D, Farber JM. Chemokine receptor responses on T cells are achieved through regulation of both receptor expression and signaling. J Immunol (1999) 162:3840–50. [PubMed] [Google Scholar]
- 50.Hakim S, Bertucci MC, Conduit SE, Vuong DL, Mitchell CA. Inositol polyphosphate phosphatases in human disease. Curr Top Microbiol Immunol (2012) 362:247–314. 10.1007/978-94-007-5025-8_12 [DOI] [PubMed] [Google Scholar]
- 51.Trinchieri G, Sher A. Cooperation of toll-like receptor signals in innate immune defence. Nat Rev Immunol (2007) 7:179–90. 10.1038/nri2038 [DOI] [PubMed] [Google Scholar]
- 52.Beers DR, Henkel JS, Zhao W, Wang J, Appel SH. CD4+ T cells support glial neuroprotection, slow disease progression, and modify glial morphology in an animal model of inherited ALS. Proc Natl Acad Sci U S A (2008) 105:15558–63. 10.1073/pnas.0807419105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tada S, Okuno T, Yasui T, Nakatsuji Y, Sugimoto T, Kikutani H, et al. Deleterious effects of lymphocytes at the early stage of neurodegeneration in an animal model of amyotrophic lateral sclerosis. J Neuroinflammation (2011) 8:19. 10.1186/1742-2094-8-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lincecum JM, Vieira FG, Wang MZ, Thompson K, De Zutter GS, Kidd J, et al. From transcriptome analysis to therapeutic anti-CD40L treatment in the SOD1 model of amyotrophic lateral sclerosis. Nat Genet (2010) 42:392–9. 10.1038/ng.557 [DOI] [PubMed] [Google Scholar]
- 55.Staats KA, Humblet-Baron S, Bento-Abreu A, Scheveneels W, Nikolaou A, Deckers K, et al. Genetic ablation of IP3 receptor 2 increases cytokines and decreases survival of SOD1G93A mice. Hum Mol Genet (2016) 25:3491–9. 10.1093/hmg/ddw190 [DOI] [PMC free article] [PubMed] [Google Scholar]