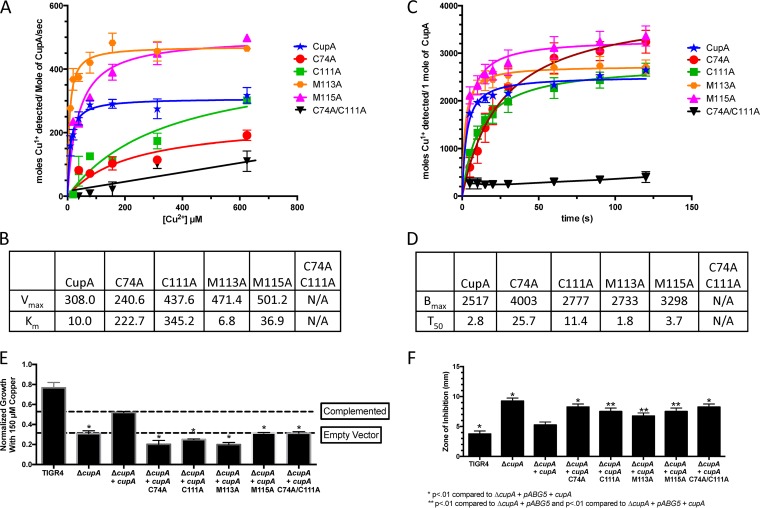
FIG 5 .
CupA’s ability to reduce Cu2+ is necessary to prevent copper stress. (A) Maximum rate of CupA Cu2+ reduction and release of Cu1+, as determined by BCS, for recombinant CupA and various mutations of the Cu1+-binding domain, as described by Fu et al. (14). Each error bar represents the standard error of the mean of three experiments. The rate was averaged over a 5-s interval. (B) Vmax and Km of wild-type CupA and variants as determined by curve fits to a Michaelis-Menten model. (C) CopY Cu2+ reduction and release of Cu1+ over time, as determined by BCS, for recombinant CupA and various mutations of the Cu1+-binding domain at a 312.5 µM Cu2+ concentration. Each error bar represents the standard error of the mean of three experiments. (D) Bmax and T50 of wild-type and mutant forms of CupA as determined by curve fits to a hyperbola one-site model fit. (E) Endpoint growth of S. pneumoniae TIGR4, the ΔcupA mutant, the ΔcupA mutant with the empty pABG5 vector, and the ΔcupA mutant complemented with wild-type and various mutant forms of cupA at 150 µM Cu2+ in THYB at pH 6.5 at 37°C. *, P < 0.01 compared to the ΔcupA mutant complemented with wild-type cupA on the pABG5 vector in a Student t test. Each strain, except TIGR4, contains pABG5. Data are representative of three independent experiments with three replicates. (F) Zone of inhibition of S. pneumoniae TIGR4, the ΔcupA mutant, the ΔcupA mutant with the empty pABG5 vector, and the ΔcupA mutant complemented with wild-type and various mutant forms of cupA on TSA blood agar plates incubated overnight at 37°C. Zones were measured as the distance from the edge of the disc containing 10 µl of 1 M Cu2+. *, P < 0.01 compared to the ΔcupA mutant complemented with wild-type cupA on the pABG5 vector; **, P < 0.01 compared to the ΔcupA mutant complemented with wild-type cupA on the pABG5 vector and the ΔcupA mutant with the empty pABG5 vector in a Student t test. Data are representative of three independent experiments with six replicates each. N/A, not available.