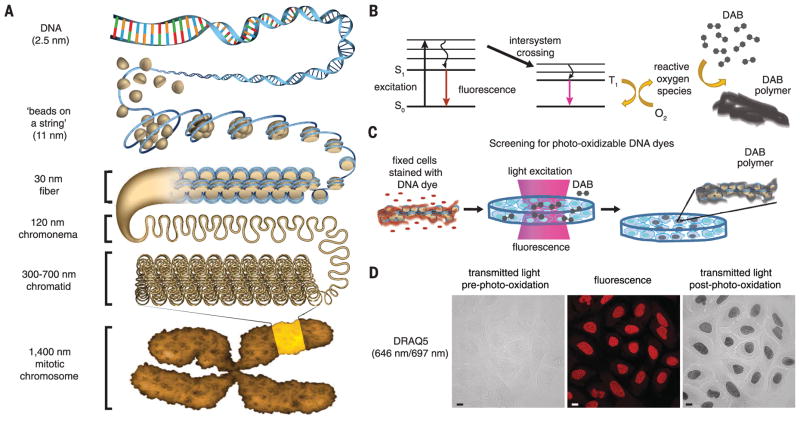
Fig. 1. A fluorescent DNA-binding dye that catalyzes local DAB polymerization on chromatin in the nucleus.
(A) Hierarchical chromatin-folding model. (B) Excited fluorophores that undergo intersystem crossing generate reactive oxygen species that catalyze DAB polymerization. S0, ground state; S1, excited singlet state; T1, excited triplet state. (C) Schema for cell-based screen for DNA-binding dyes that photo-oxidize DAB. (D) U2OS cells were fixed with glutaraldehyde and stained with DRAQ5. Cells were incubated with DAB and excited by continuous epifluorescence illumination for 5 min. DAB photo-oxidation was identified by the appearance of dark DAB precipitates in the nucleus. Fluorescence (middle), transmitted-light images pre– (left panel) and post–photo-oxidation (right panel). Scale bar, 10 μm. See Movie 1 for photo-oxidation of DAB by DRAQ5.