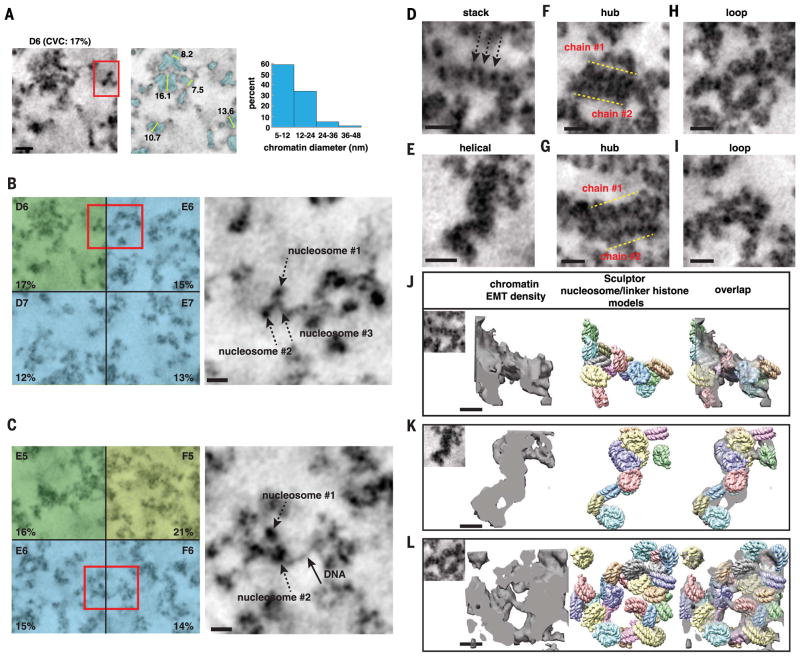
Fig. 5. DNA and nucleosomes form disordered chromatin chains that have different particle arrangements, conformations, densities, and 3D motifs.
(A) In interphase subvolumes that have low CVCs (<20%), chromatin chains have predominantly 5- to 12-nm diameter bin distributions (see fig. S5). Left, single TS (D6, SAEC #1); middle, segmented chromatin; right, chromatin-diameter range. Scale bar, 20 nm. (B to C) Magnified images of chromatin in volumes with low CVCs (red boxes). Chromatin chains have thin threads decorated with discrete particles (right panels: single TS, scale bar 20 nm). See movies S1 and S2. (D to I) Gallery of different structures and motifs: (D) short linear nucleosome stack, (E) helical twist, (F) and (G) two chromatin chains interact in parallel to form a hub, and (H) and (I) loops between and within chromatin chains. Scale bar, 20 nm. (J to L) Sculptor models (middle) of nucleosome–linker histone H5 (PDB 4QLC) in EMT densities of chromatin stack, helical twist, and loop (left panels). Overlap of the Sculptor models and EMT densities (right). One Sculptor solution is shown. Cross-correlation values: 82% (J), 92% (K), and 86% (L). Additional Sculptor solutions in fig. S11. Scale bar, 11 nm.