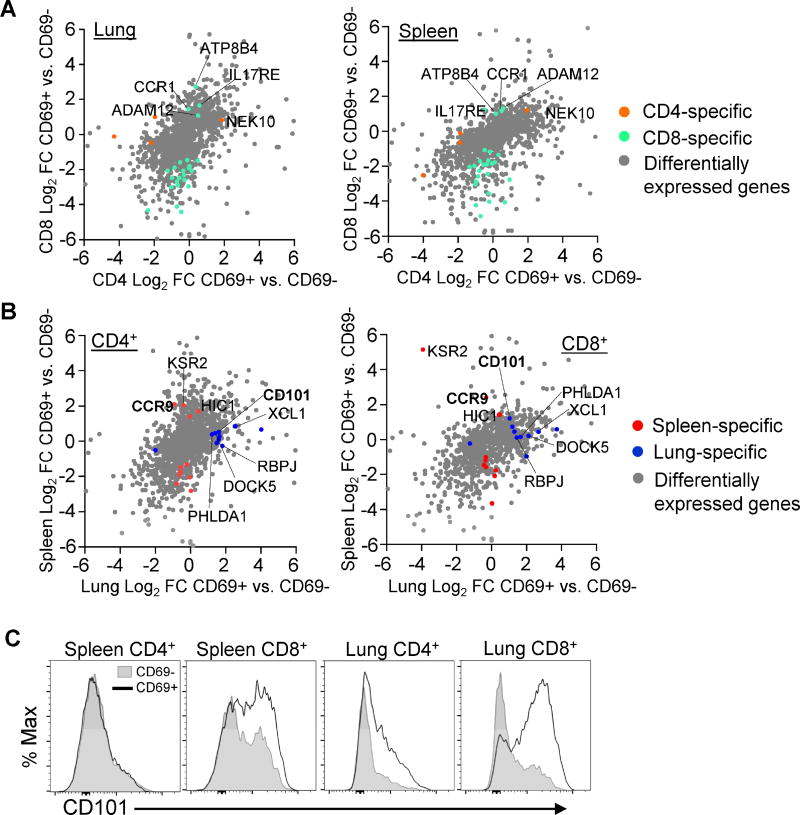
Figure 6. Lineage and tissue-specific transcription and phenotypic profiles in human CD69+ memory T cells.
(A) Analysis of lineage-specific gene expression in CD69+ and CD69− memory T cells. Scatter plots display log2 fold change of CD4+CD69+ vs. CD69− on the x axis and CD8+ subsets on the y axis from lung (left) and spleen (right). Grey dots represent genes with significant differential expression in any paired CD69+ vs. CD69− sample. Orange dots (“CD4 specific”) represent genes with significant differential expression in CD4+ CD69+ vs. CD69− but not in CD8+ samples. Green dots (“CD8 specific”) represent genes with significant differential expression in CD8+CD69+ vs. CD69− but not in CD4+ samples. (B) Analysis of tissue-specific genes in CD69+ and CD69− memory T cells. Scatter plots display log2 fold change of lung CD69+ vs. CD69− samples on the x axis and spleen samples on the y axis for CD4+ (left) and CD8+ (right) T cells using the same strategy as in (A) with red dots denoting “spleen specific” and blue dots denoting “lung specific” transcripts in the paired analysis. (C) CD101 expression in human tissues. Representative plots show CD101 expression in CD69+ (black outline) and CD69− (shaded) cells from one individual donor. Data are representative of 15 donors (see Figure S5).