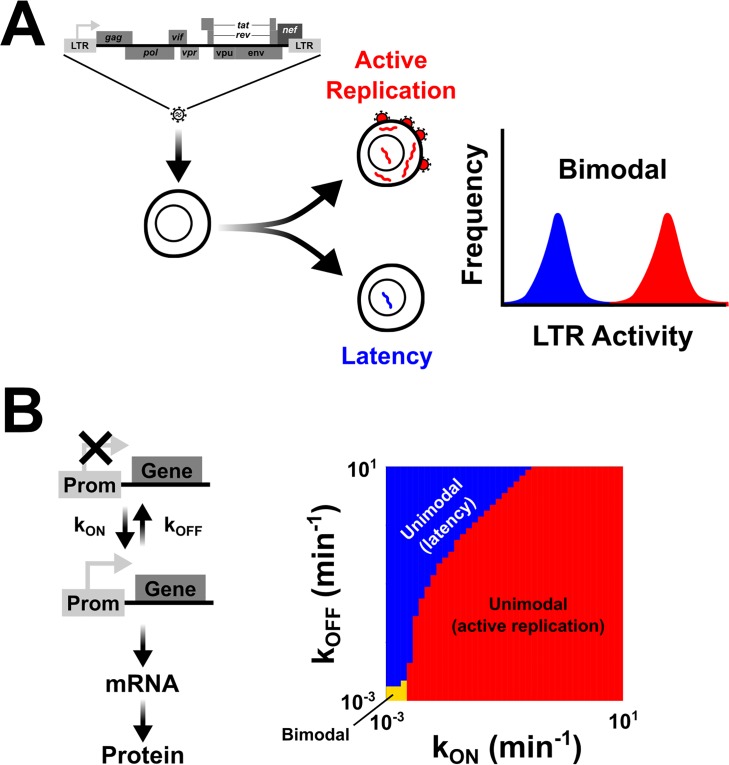
Fig 1. The mechanistic problem underlying bimodal fate-selection programs: Promoter toggling is theoretically sufficient to generate bimodality, but only in a narrow parameter regime.
(A) A simplified fate-selection decision in HIV. Upon infection of a CD4+ T lymphocyte, HIV either enters into an active state of replication (red), producing viral progeny and destroying the host cell, or enters into a quiescent state of silenced gene expression termed proviral latency (blue). This fate bifurcation between active replication and latency is not controlled by the cell state [27] but rather by an HIV gene-regulatory program that can generate bimodal gene-expression distributions from its long terminal repeat (LTR) promoter. (B) The LTR is accurately described by a 2-state promoter model (e.g., random telegraph models) in which the LTR switches between an inactive (represented by Prom-Gene that is crossed out, top) to an active (represented by Prom-Gene) state of expression at rate kON. In the active state, mRNAs are produced, before the promoter flips back to the inactive state at rate kOFF. Promoter toggling between these active and inactive states can produce bimodal distributions in gene-expression products, but only within a restricted regime of phase space. Each parameter set was checked to see if it generated unimodal latency (blue), unimodal active replication (red), or bimodality (orange), as described in the Materials and methods section. For the modality analysis, each mode was required to contain at least 0.1% of the population; otherwise, the parameter set was determined to produce a unimodal population.