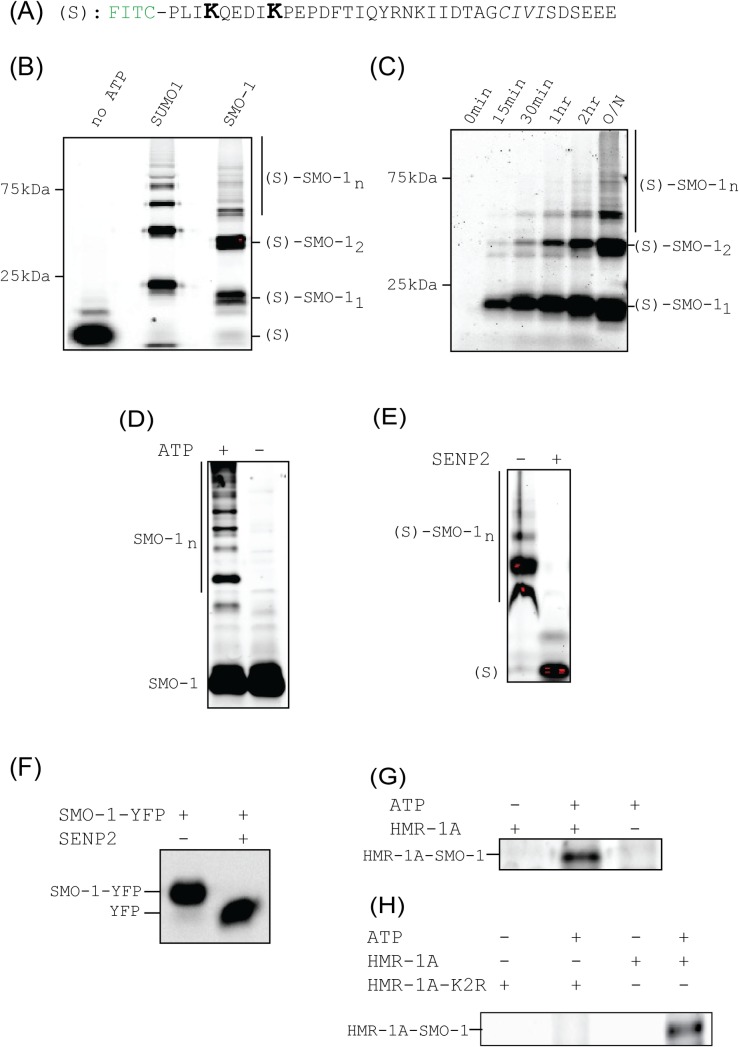
Fig 6. SMO-1 forms chains in in vitro SUMOylation assays.
(A) Sequence of FITC fluorophore labelled peptide used as substrate in in vitro SUMOylation assays. The lysines competent for SUMOylation are in bigger font and in bold face. All subsequent images were obtained by observing FITC fluorescence at 519 nm unless otherwise mentioned. (B) SUMOylation reaction products with peptide shown in (A) and SUMO1/SMO-1 resolved on SDS-PAGE gel. The negative control is lane labelled “no ATP”. Bands of free peptide and peptide conjugated with one, two or multiple (n) SMO-1 are marked. The molecular weight marker positions are indicated. (C) Time course of SUMOylation reaction between peptide shown in (A) and SMO-1. Different time points are indicated. O/N means overnight (~12 hours) incubation of reaction. Band identities and molecular weight marker positions are shown as in (B). (D) SUMOylation reaction using FITC-labelled SMO-1 and in absence of any other peptide substrate. The reaction products and the negative control (without ATP) are resolved and marked. (E) De-conjugation of SUMOylated peptides by SENP2 enzyme. The SUMOylation reaction products shown in (B) were run with or without treatment with SENP2. (F) De-conjugation reaction of SMO-1-YFP construct with SENP2. The SDS-PAGE gel is imaged by observing YFP fluorescence. (G) SUMOylation reaction between FITC-labelled SMO-1 and HMR-1A, a known protein substrate of SMO-1 in C. elegans. The reaction product band is marked. Negative controls–without ATP and without HMR-1A is also shown. (H) SUMOylation reaction described in (G) is shown with the negative control where all lysines of HMR-1A were mutated to arginines to create the SUMOylation incompetent HMR-1A-K2R construct.