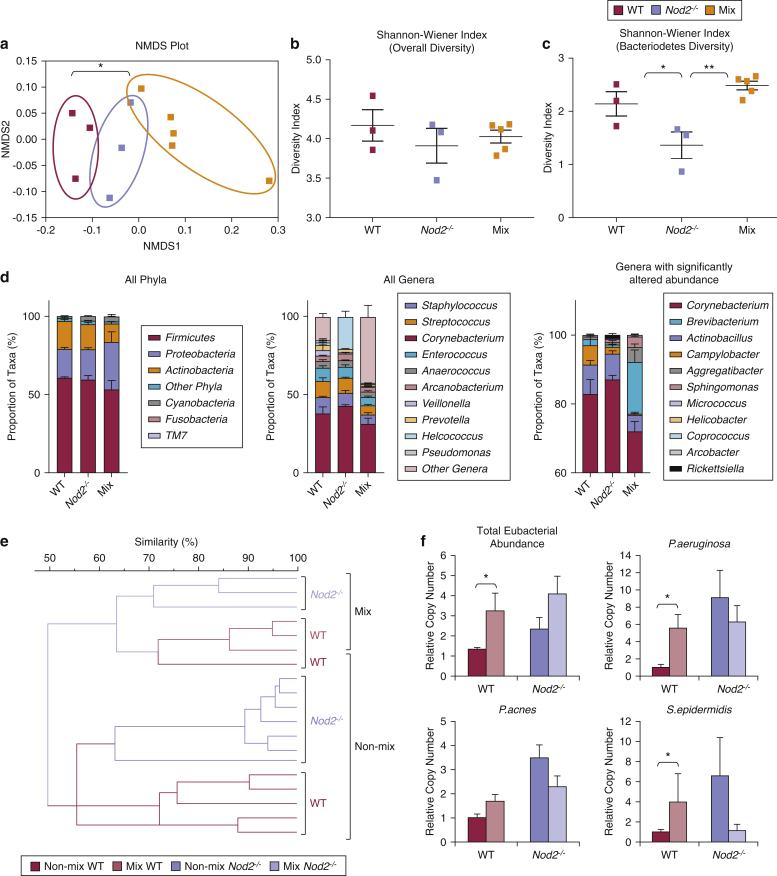
Figure 5.
WT mice co-housed with Nod2–/– mice from birth acquire pathogenic bacteria. WT and Nod2–/– litters were mixed from birth and wounded in adulthood, and their wound microbial communities sequenced. (a) NMDS plot showing differences in clustering of microbial communities. Alpha diversity of wound tissue across (b) all microbial communities and (c) the Bacteriodetes phylum was compared using the Shannon-Wiener index. (d) Taxonomic classification of the skin microbiome showing proportions of bacteria in each treatment group at the phylum level and genus level and genera that were significantly altered between treatment groups. Individual taxa with abundances too low to visualize clearly and unassigned reads are grouped into the “other” category, which comprises eight additional phyla plus unassigned reads at the phylum level and 219 additional genera plus unassigned reads at the genus level. (e) UPGMA dendrogram of WT and Nod2–/– wound tissue DGGE fingerprints. (f) Total wound eubacterial abundance (16S real-time PCR) was significantly increased in WT mice co-housed with Nod2–/– mice. Mean + standard error of the mean. All data are representative of two independent experiments with n = 3 mice/non-mix groups and n = 5 mice/mix group. ∗∗P < 0.001, ∗P < 0.05. NMDS, non-metric multidimensional scaling; UPGMA, unweighted pair group method with arithmetic mean.