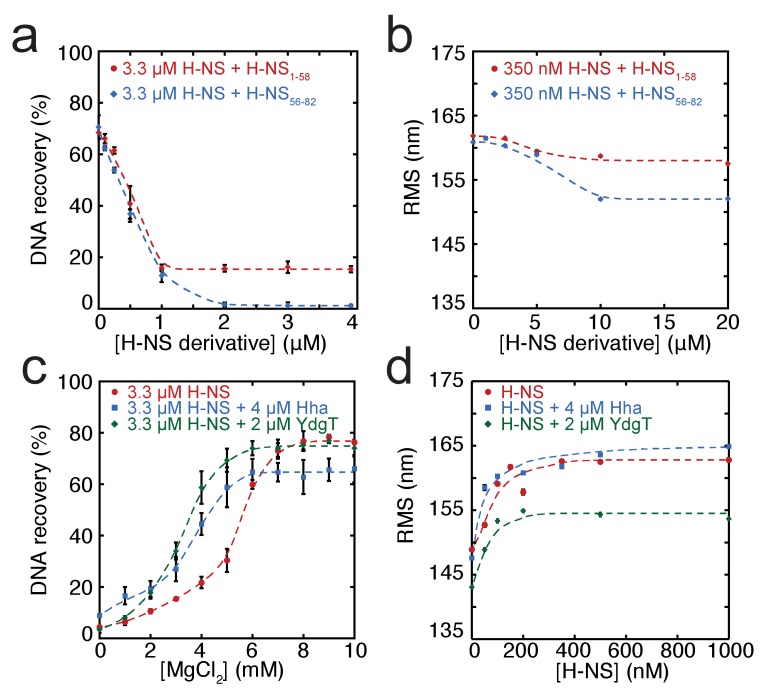
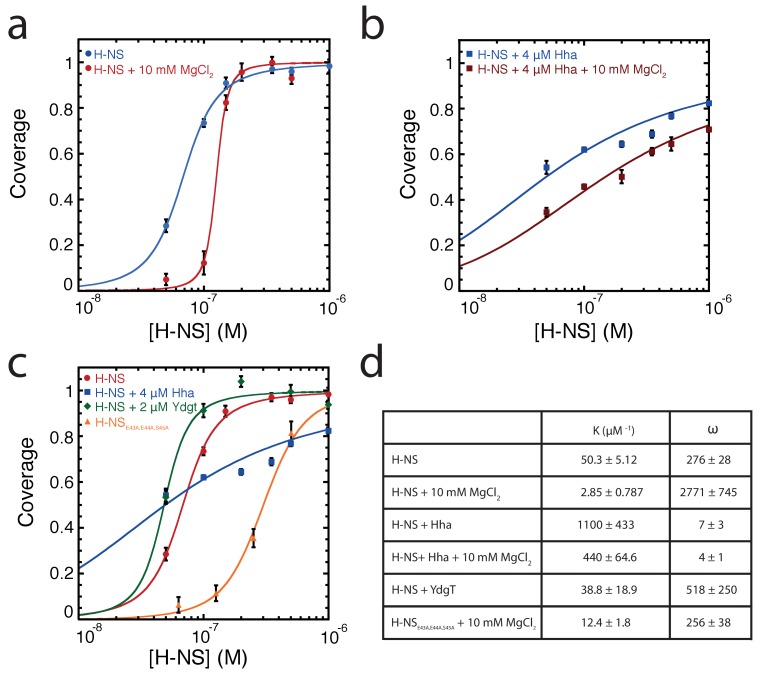
Figure 3. Modulation of H-NS function by protein cofactors.
(a) DNA bridging efficiency as a function of inhibiting peptides targeting either the dimerization domain (H-NS1-58) and multimerization domain (H-NS56-82). (b) Root Mean Square displacement (RMS) as a function of inhibiting peptides targeting either the dimerization (H-NS1-58) and multimerization (H-NS56-82) (N > 70, for each point). (c) DNA bridging efficiency as a function of Mg2+ concentration in the presence and absence of 4 µM Hha or 2 µM YdgT. (d) RMS of DNA in the presence of H-NS, H-NS-Hha, and H-NS-YdgT. Dashed lines are to guide the eye (N > 60, for each point).