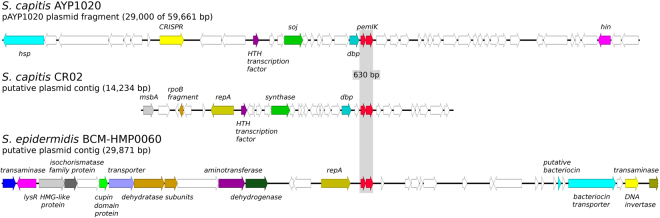
Figure 7.
The location of the pemIK-Scap operon in S. capitis and S. epidermidis. The operon was found in three completely different contexts in either a plasmid sequence of pAYP1020 or a putative plasmid sequence. White arrows indicate sequences that encode hypothetical proteins, and coloured arrows indicate the following genes in S. capitis AYP1020: heat shock protein 1, hsp, cyan; CRIPSR-related DNA-binding protein, yellow; putative HTH transcription factor, purple; sporulation initiation inhibitor protein, soj, green; putative DNA-binding protein, sky blue; pemIK locus, red; and DNA invertase, hin, magenta. In S. capitis CR02: lipid A export ATP-binding/permease protein MsbA fragment, msbA, light grey; rpoB fragment, beige; replication protein A, repA, olive; putative HTH transcription factor, purple; cobyrinic acid a,c-diamide synthase, green; putative DNA-binding protein, sky blue; and pemIK locus, red. In S. epidermidis BCM-HMP0060: histidinol-phosphate transaminase family protein, blue; LysR family transcriptional regulator, magenta; HMG-like protein, light grey; isochorismatase family protein, dark grey; cupin domain protein, light green; putative transporter, pale purple; leuC, leuD, 3-isopropylmalate dehydratase subunits, beige; class I/II aminotransferase, dark purple; type I glyceraldehyde-3-phosphate dehydrogenase, gap, dark green; replication protein A, repA, olive; pemIK locus, red; putative bacteriocin and bacteriocin transporter, cyan; putative transposon DNA-invertase Bin3, yellow; and putative histidinol-phosphate transaminase, dark olive.