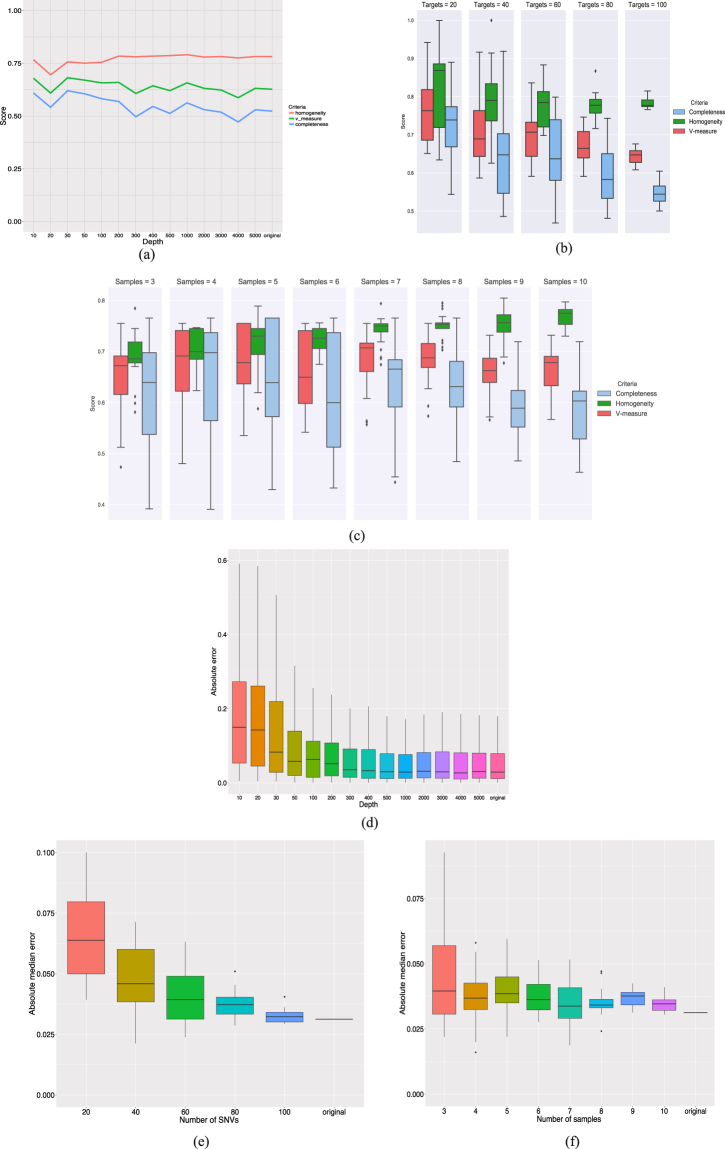
Figure 5.
Results of simulation studies for Experiment 2 (aneuploid cell lines DAH55 and DAH56). (a) Experiment 2, V-measure, downsampling read depth. The vertical axis indicates the V-measure, homogeneity and completeness scores obtained by applying PyClone for each read depth considered when downsampling the number of variant and reference reads aligned to each SNV. (b) Experiment 2, V-measure, downsampling the number of targets. Box plots of the V-measure, homogeneity and completeness scores obtained by applying PyClone to each simulated data set under each number of targeted SNVs considered. (c) Experiment 2, V-measure, downsampling the number of samples. Box plots of the V-measure, homogeneity and completeness scores obtained by applying PyClone to each possible number of samples considered. (d) Experiment 2, prevalence error, downsampling read depth. Box plots of the absolute errors in estimating cellular prevalence obtained by applying PyClone to each read depth considered. Note that outliers were omitted from the box plots to facilitate visualization of the median, first and third absolute error quartiles. (e) Experiment 2, prevalence error, downsampling number of targets. Box plots of the median absolute errors obtained by applying PyClone to each simulated data set under each number of targeted SNVs considered. (f) Experiment 2, prevalence error, downsampling number of samples. Box plots of the median absolute errors obtained by applying PyClone to each simulated data set under each number of samples considered.