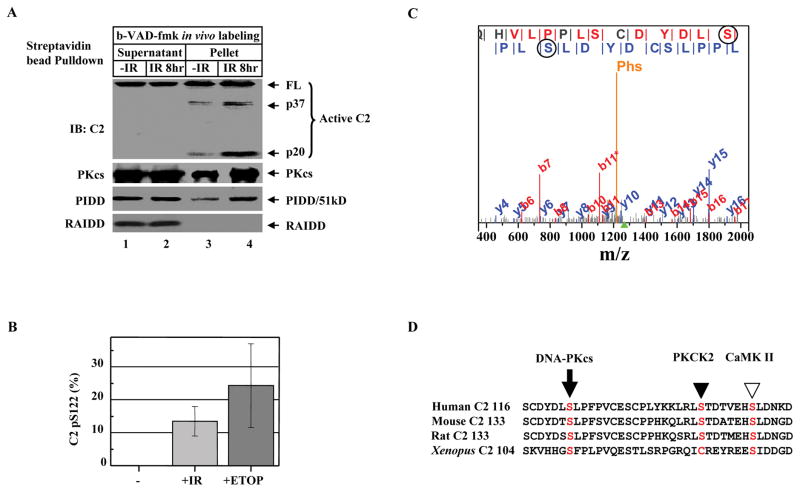
Figure 2. DNA Damage Induces Caspase-2 Serine 122 Phosphorylation.
(A)DNA damage activates caspase-2 bound to DNA-PKcs-PIDDosome. HeLa cells were exposed to 80 Gy of ionizing radiation (IR). Eight hrs after IR, cells (2X107) were labeled with b-VAD-fmk (4 μM) for 1 hr, nuclear extracts were subjected to streptavidin bead-pulldown, and immunoprecipitates were analyzed by IB against the molecules indicated.
(B) Quantification of phospho-peptides containing phosphorylated S122 (pS122). HEK293 cells stably expressing FLAG-tagged caspase-2 (C303A) were left untreated (-), exposed to 80 Gy IR (+IR), or treated with 100 μM etoposide (+ETOP) for 2 hrs. Caspase-2 was FLAG-affinity-purified, and phosphorylation of caspase-2 was analyzed by MudPIT combined with spectral count analysis. Each bar represents the mean ±S.D. of three independent experiments (n=3).
(C) Annotation of a representative tandem mass spectrum matched to L.SGLQHVLPPLSCDYDLpSLPFPV.C from elastase-digested caspase-2. The doubly charged precursor ion with a protonated mass of 2535.71 Dalton is marked by the green triangle under the m/z-axis, while the phosphate neutral loss peak is denoted with “Phs”. Fragment ions bearing phosphorylated serine 122 (circled) are matched in both b and y ion series, which are labeled in red and blue, respectively.
(D)Sequence alignment of human caspase-2 (residues 116–153) with mouse, rat, and Xenopus caspase-2 showing the conserved DNA-PKcs phosphorylation site S122 (arrow). The PKCK2 phosphorylation site S157 in human caspase-2 (solid arrowhead) and the CaMKII phosphorylation site S135 in Xenopus caspase-2 (open arrowhead) are also shown.