Abstract
Protein biosensors are widely used for the monitoring of metabolite concentration and enzymatic activities inside living cells and in in vitro applications. Neutrophil elastase (NE) is a serine protease of relevance in inflammatory diseases whose activity can lead to pathological conditions if unregulated. This study focuses on the structural characterization of a biosensor for NE activity based on Förster resonance energy transfer (FRET). The cleavage by NE results in dissociation of the FRET fluorescent protein pair and alteration of the fluorescent emission spectrum. We have used small angle x-ray scattering at a high intensity synchrotron source, combined with model-free analysis of the scattering data, to demonstrate the structure of the biosensor and the effect of its exposure to NE on size and shape. These investigations, together with biochemical studies, established the nanostructure-activity relationship that may contribute to the detailed understanding of the FRET-based biosensor and guide the rational design of new biosensor constructs.
Introduction
Fluorescent protein biosensors are powerful bioanalytical tools applied to the measurement of biomarkers in vitro and to the monitoring of biological processes in living cells (1). Protein-based biosensors are mostly formed by a fluorescent module and a target-specific recognition module (2, 3). On target recognition, a conformational change of the protein induces an optical response. The green fluorescent protein (GFP) and its variants are the most used fluorescent components in biosensors because of their autocatalytic maturation process, ease of expression, and conformational stability (4). The recognition module can have different sizes and structure, ranging from single reactive residues such as the cysteines in a hydrogen peroxide biosensor (5), to peptides such as in metal-sensing biosensors (6), to whole proteins such as in sugar-binding biosensors (7). When two fluorescent proteins are in close proximity and their emission and excitation spectra overlap at least partially, Förster resonance energy transfer (FRET), which is the radiationless transfer of energy between two fluorescent molecules, can occur. FRET-based biosensors produce a ratiometric signal that, calculated as the ratio between the emission at two wavelengths, is independent of the concentration of the biosensor in the sample (8). They are preferred to single fluorescent proteins as the latter provide only a read-out as fluorescence intensity. This aspect is especially important when the biosensors are recombinantly expressed in living cells and cell compartments. By selecting reporter proteins with different fluorescent properties and by engineering the recognition module, a variety of protein biosensors has been developed able to specifically monitor minor changes of ion and metabolite concentration (7, 9, 10, 11, 12), of the redox status (13), and of enzymatic activities (10, 14) in various biological systems. The conformational changes in the protein biosensors that cause their response to external stimuli are usually analyzed by fluorescence spectroscopy in solution, or by fluorescence microscopy when expressed in living cells (15, 16). However, despite the fact that the nanostructure of the FRET-based protein biosensors is decisive for the FRET effect and thus the activity of the sensor, the related literature is still limited.
Here, we structurally characterize a FRET biosensor for neutrophil elastase (NE), a serine protease of relevance in inflammatory diseases and whose activity can lead to pathological conditions if unregulated (17). In our NE biosensor, the yellow fluorescent protein (YFP) and the cyan fluorescent protein (CFP) are linked via a recognition sequence specific for human NE (18). When the biosensor is intact, FRET occurs and the excitation of the donor (CFP) results in an enhanced emission of the acceptor molecule (YFP) (17). In the presence of NE, the dissociation of the fluorescent moieties results in a spectral response (Fig. 1). To the best of our knowledge, the solution and crystal structure of both the designed biosensor construct and modified fluorescent proteins are not available in literature. X-ray crystallography and most electron microscopy methods only allow the study of protein conformation in a crystalline state (19). The determination of protein conformation in a native, functional state in solution is currently only possible with small angle x-ray scattering (SAXS), cryogenic electron microscopy, or NMR spectroscopy, with the latter method limited to small molecules and high concentration (19, 20). In our study, SAXS was applied to investigate the solution structure of the designed biosensor, with a focus on orientation of the two linked proteins in the tandem construct at a nanometer scale resolution. This method can further detect both ordered and disordered regions within a protein population (21). In a recent study, this method was used to research the conformational changes and the flexibility of single-molecule FRET biosensors to monitor phosphorylation and calcium presence (22). Here we present, to our knowledge, the first study combining biochemical methods with spectroscopy, synchrotron SAXS, and modeling of the scattering data using model-free approaches to correlate nanostructure and function to the site-specific cleavage of the FRET protein pair with NE.
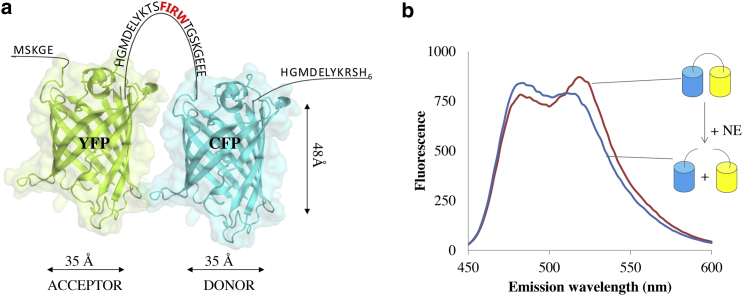
Figure 1.
(a) Schematic view of the biosensor for NE activity. The biosensor is formed by the YFP-CFP FRET pair connected by a linker peptide carrying the NE-specific recognition sequence Phe-Ile-Arg-Trp. The GFP variants YFP and CFP are represented using the available three-dimensional structure of the wild-type GFP (PDB: 1EME). (b) Fluorescence emission spectra of the biosensor in the intact and cleaved form obtained by incubation with NE. To see this figure in color, go online.
Materials and Methods
Overexpression and purification of the biosensor
An aliquot of 2 μL of a glycerol stock of the chemically competent Escherichia coli BL21(DE3)GOLD cells previously transformed with the pET22b(+)-CFP-NE1-YFP plasmid was used to inoculate a 100 mL culture containing lysogeny broth medium supplemented with 50 μg/mL ampicillin (95% purity; Sigma-Aldrich, Buchs, Switzerland). After overnight cultivation at 37°C at 180 rpm, this was diluted to 500 mL of selective medium to a starting OD600 = 0.05, and incubated at 37°C until it reached an OD600 = 0.6–0.8. Gene expression was induced with 1 mM isopropyl β-D-thiogalactoside (purity ≥99%; Sigma-Aldrich) and the temperature lowered to 18°C. Cells were harvested after 16–20 h incubation by centrifugation and the cell pellet was resuspended in 100 mM potassium phosphate buffer (pH 7.4; 5 mL/g cells) and treated with 1 mg/mL lysozyme (62971–10G-F; Sigma-Aldrich) and a protease inhibitor mix (one tablet per 50 mL suspension, cOmplete Roche Complete Protease Inhibitor Mix, EDTA-free obtained from Sigma-Aldrich). Cells were incubated 30 min on ice before being frozen at −80°C for 30 min. Cells were then treated with benzonase nuclease (1 μL/2 g cells; New England Biolabs Inc., Ipswich, MA) and incubated at 37°C for 1 h under gentle shaking. Cell disruption was performed by sonication with a Branson sonicator (Branson Ultrasonics) and the cell debris was removed by ultracentrifugation (47,808 × g, 4°C, 40 min, SS34 rotor; Sorvall). As the biosensor carries a hexa-histidine tag, purification proceeded by affinity chromatography by fast protein liquid chromatography (Äkta Purifer; GE Healthcare, Little Chalfont, United Kingdom) using a HisTrap HP column (volume = 5 mL; GE Healthcare) preequilibrated with 100 mM potassium phosphate buffer (pH 7.0) and 10 mM imidazole (p.a. grade; Sigma-Aldrich). Proteins were eluted with a 10–500 mM imidazole linear gradient, and fractions containing the biosensor were pooled and buffer-exchanged to 100 mM potassium phosphate buffer (pH 7.0). The biosensor was further purified by size exclusion chromatography using a Superdex 75 10/300 GL (GE Healthcare) column in phosphate-buffered saline (PBS; pH 6.2) containing 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 (all p.a. grade; Sigma-Aldrich). Protein purity was assessed by analyzing 5–10 μg of biosensor by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (10% acrylamide resolving gel, 5% stacking gel) and the protein concentration was calculated using the extinction coefficient calculated in silico (17).
Cleaving of the biosensor
The cleaved version of the biosensor (concentration = 0.58 mg/mL) was prepared by incubating the biosensor in the presence of NE (1 mU/mL, final concentration), at a biosensors/NE molar ratio of ∼100, in PBS buffer (pH 6.2) for 3 h at room temperature. Buffers were filtered using a 0.22 μm filter and samples were stored at −20°C until measurement.
Spectroscopic characterization
Fluorescence measurements were performed with a multiwall-plate reader (Varian, Palo Alto, CA) in black, 96-well, half-area plates using 60 μL samples at a protein concentration of 0.58 mg/mL, with λex = 395 nm and λem = 460–660 nm.
Dynamic light scattering
Dynamic light scattering (DLS) was performed on a Malvern Zetasizer Nano ZS90 (Malvern Instruments, Westborough, MA) at 25°C with a He−Ne Laser beam, at a wavelength of 633 nm, laser power of 4 mW, and a scattering angle of 90°. The protein construct was measured in PBS (pH 6.2). The mean hydrodynamic radius (Rh) and the size distribution (polydispersity index) of the biosensor were determined with the cumulant method.
SAXS
SAXS measurements were performed at the Austrian SAXS beamline at ELETTRA, Trieste, Italy (23). The samples (protein concentration = 0.58 mg/mL in PBS buffer (pH 6.2)) were measured in a 1.5-mm diameter quartz capillary mounted in the x-ray beam at 25°C. An x-ray beam with a wavelength of 0.77 Å (16 keV) was used, with a sample to detector distance of 1830 mm providing a q-range from 0.1 < q < 7 nm−1, where q is the length of the scattering vector, defined by q = 4π/λ sin(θ/2), λ being the wavelength and θ being the scattering angle. The two-dimensional SAXS patterns were acquired for 27 s, with 3 s delay between frames, using a Pilatus3 1M detector (Dectris, Baden, Switzerland; active area of 169 × 179 mm2 with a pixel size of 172 μm). The two-dimensional SAXS pattern were integrated into one-dimensional scattering function I(q) using Fit2D (24), and then analyzed with IGOR pro (Wavemetrics, Lake Oswego, OR).
Measurements were done in triplicate to check for beam damage, and the average of the results was used. No beam damage was observed. The scattering of PBS buffer was subtracted from the measurements as background.
Analysis of SAXS data
Guinier analysis, from the Guinier approximation of the scattering data and further analysis of the calculated pair distance distribution function p(r), (see below), was performed to calculate the radius of gyration (Rg).
For particles of arbitrary shape with an electron density difference of Δρ(r) relative to the mean value, the p(r) is given by p(r) = r2Δρ̃2(r), where Δρ̃2(r) is the convolution square of the electron density averaged for all directions in space. This averaging causes no loss of information in the case of particles with spherical symmetry. The indirect Fourier transformation method was used for a model-free analysis of the scattering data (25, 26, 27).
The p(r) is calculated from the scattered intensity I(q) using the following equation (26):
| (1) |
giving a real-space representation of the overall shape of the proteins in case of negligible interparticle-interactions at the low concentrations in this study (27). The radius of gyration was also calculated from the p(r) using
| (2) |
A low-resolution, three-dimensional model of the proteins was also reconstructed from the SAXS data using the automated bead modeling approach in Dummy Atom Model Minimization (DAMMIM) (28, 29). The shape reconstruction was started from densely packed beads inside a spherical search volume with Dmax approximated from the p(r). Ten independent DAMMIM runs were performed. The most divergent models were discarded and resulting models averaged (30).
The Kratky plot (q2I(q) versus qRg) was used to study protein folding and symmetry (31).
Results and Discussion
The YFP-CFP protein pair and its evolved variants have become one of the most used fluorescence protein FRET pairs to investigate protein-protein interactions within the cell, and to study kinase and phosphatase activities (32, 33, 34). The response of the sensor was mostly monitored by fluorescence spectroscopy methods without, however, providing a detailed structural insight into the construct (35). By complementing the fluorescence-based measurements with SAXS analyses, a better understanding of both the FRET effect and the orientation of the proteins in the tandem FRET pair, responsible for this effect, can be achieved.
The YFP-CFP protein pair in this study is connected by a peptide carrying the recognition sequence Phe-Ile-Arg-Trp, specific for human NE (see Fig. S1 for the primary and secondary structure). It is the biosensor carrying the shortest linker sequence among the ones previously reported in literature (17). Upon incubation with NE, hydrolysis of the peptide bond in the linker sequence results in dissociation of the fluorescent moieties and a spectral response (Fig. 1). The FRET ratio, calculated by dividing the emission at 520 nm and 485 nm, is thus reduced from 1.1 to 0.8 on cleavage under simulated physiological conditions (17).
SDS-PAGE analysis shows that the ∼60 kDa biosensor is cleaved into ∼33 kDa GFP-like subunits (Fig. S2). According to the structure and location of the NE recognition site, cleavage results in the dissociation of the two folded CFP and YFP barrel subunits carrying additional unstructured stretches (Fig. S1). YFP carries a five-amino-acid long, short N-terminus stretch with an estimated length of ∼1.7 nm and partial linker region of ∼4.7 nm, and CFP carries the partial linker region of ∼4.3 nm and the 15-amino-acid long, C-terminus tail containing the His-tag that is ∼6.0 nm long.
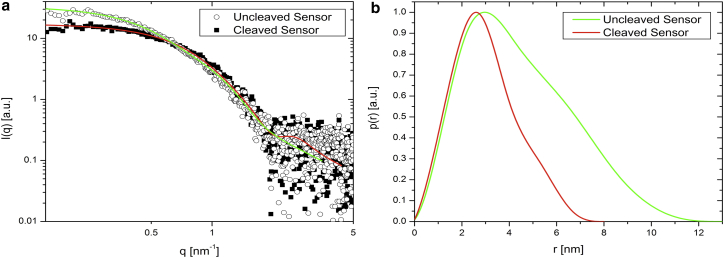
The SAXS I(q) profiles for the biosensor before and after reaction with NE, in combination with the model-independent fits calculated with the indirect Fourier transformation method (26, 27), using Eq. 1, with the corresponding p(r) functions, are presented in Fig. 2, a and b, respectively. The decrease in forward scattering intensity in the I(q) profiles between the uncleaved to the cleaved biosensor is characteristic for a decrease in size of the nanoobject. This region of the scattering curve is sensitive to aggregation, with a theoretical intensity difference of a factor of two between monomers and dimers. On cleaving the biosensor, the ratio between the forward scattering intensities in Fig. 2 decrease by a factor of ∼1.8, hence we estimate that around 90% of the biosensor was cleaved during the reaction with NE.
Figure 2.
(a) Experimental SAXS curves (symbols) and calculated fits (full lines) of the biosensor before and after cleaving. (b) The corresponding p(r) functions calculated from (a) using Eq. 1. To see this figure in color, go online.
The shape of the calculated p(r) functions from SAXS for this biosensor with two broad humps indicates two linked proteins. The linear decay of the p(r) at higher r values is characteristic for ellipsoidal shape with the maximum dimension of the construct at p(r) = 0 at ∼12 nm (36). The shape of the p(r) function for the cleaved biosensor indicates elongated proteins with less assymetry and a maximum dimension at p(r) = 0 at ∼8 nm.
As it is theoretically not possible to simultaneously deduce the geometrical shape and the size distribution of a polydisperse system of particles from SAXS data, we used SDS-PAGE and DLS measurements for information on size and polydispersity of the biosensor (37). SDS-PAGE confirmed the high purity of the biosensor and cleavage on treatment with NE into molecules of approximately half weight (Fig. S2). The DLS measurements of the uncleaved biosensor indicated a hydrodynamic radius of Rh ∼4.5 nm and a polydispersity index value of ∼0.30. Hence, the larger dimensions deduced from SAXS compared to the diameter obtained from DLS with 2Rh ∼9.0 nm, may result from the deviations from spherical shape, as the Rh describes the diffusion of a spherical particle with the diffusion constant of the nonspherical construct.
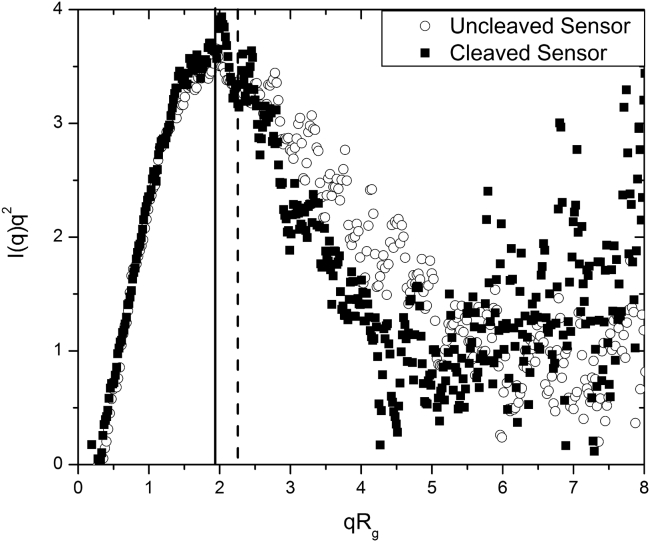
The radius of gyration (Rg), calculated from the SAXS data, confirms the change in size of the protein biosensor on reaction with NE. Rg values for the uncleaved and cleaved sensor were ∼3.1 and 2.3 nm, respectively, calculated from Guinier analysis of the experimental SAXS patterns. These results are in good agreement with the Rg values of ∼3.5 and 2.3 nm, calculated from the corresponding p(r) functions using Eq. 2. This reduction in size and transition in shape and morphology on reaction with NE does not compromise the mostly folded state of the proteins in the two constructs, as indicated by the Kratky plot in Fig. 3. The Kratky plots of both the construct and cleaved sensor exhibit a characteristic maximum, indicating mostly folded proteins. The position of the peak maximum in the Kratky plot provides further insight into the symmetry of the proteins. A theoretical maximum at qRg = is expected for globular, compact particles with deviations to larger values indicating asymmetry or flexibility in the proteins (38). The peak maximum is at qRg ∼2 for both curves in Fig. 3, suggesting an asymmetric protein shape. The maximum for the uncleaved sensor appears at slightly higher qRg values than that for the cleaved version. This indicates a more pronounced asymmetric shape of the uncleaved construct, in agreement with the results obtained from the p(r) analysis above.
Figure 3.
Kratky plot showing a characteristic pattern for mostly folded proteins of asymmetric shape for both, the uncleaved and cleaved biosensor in PBS buffer. The vertical lines indicate the maximum of the peak for the cleaved (full line) and uncleaved sensor (dashed line).
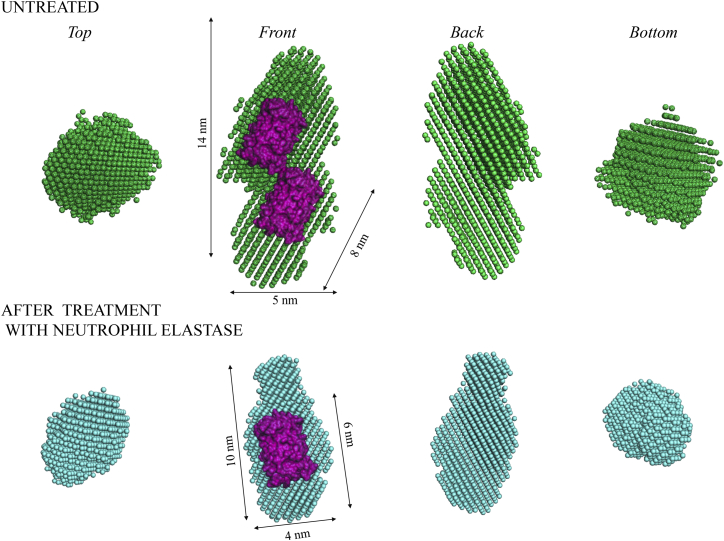
Low-resolution, three-dimensional models reconstructed from the SAXS data with the automated bead modeling approach in DAMMIM (28, 30) were used to obtain information on the orientation of the proteins in the construct, and to complement the information on the size and shape of the proteins (Fig. 4).
Figure 4.
Low-resolution three-dimensional model of the proteins calculated from the SAXS data in Fig. 2, using the automated bead modeling approach in DAMMIN. The side and top view of the uncleaved sensor (top) and the cleaved sensor (bottom) are presented together with a possible superimposition with the crystal structure of GFP subunits (PDB: 1EME). The GFP structure is shown as surface in purple and the dimensions of the ab initio model are obtained with PyMOL (The PyMOL Molecular Graphics System, version 1.8; Schrödinger). To see this figure in color, go online.
The reconstructed shape of the uncleaved sensor appears as an assembly of the two individual elongated proteins that are linked with the ∼10 nm peptide containing the NE recognition sequence. The orientation of the two proteins in the construct appears parallel, but shifted against each other along their major axis, as seen in the front and back view of the untreated construct in Fig. 4. The reconstructed shape of the individual proteins is in good agreement with the high-resolution protein structure of GFP (PDB: 1EME) (39), presented as best possible superimposition in Fig. 4. The additional modifications of the proteins with peptide segments of up to 6 nm in length, combined with hydration of the proteins, may be held responsible for differences between the constructs in solution and the superimposed protein crystal structure in vacuum. The presence of a minor portion of the uncleaved version in the cleaved sample may also contribute to deviations of the model from the crystal structure (see Results and Discussion, above).
For the uncleaved biosensor, the polydispersity in the relative alignment of the linked protein subunit contributes to the observed differences in size and shape, and might affect the functionality of the biosensor. Because the alignment of the two proteins in the construct is decisive for the FRET effect, this detailed nanostructural insight into the biosensor construct can be applied for a rational design of improved biosensor structures. For instance, the introduction of a second linker region, connecting the free ends of the two proteins, could hold them in a more compact conformation and reduce the polydispersity in the alignment of the two proteins in the tandem construct. This may improve the FRET and hamper possible protein denaturation processes. In a follow-up study, we also plan to further optimize the alignment of the fluorescent proteins through systematic variation of the length and composition of the linker peptide, as well as engineer the amino acid sequence of the interprotein region. Achieving optimized FRET would lead to more effective sensors with better spectroscopic response and detection limit for NE. We further intend to entrap the sensor in lyotropic liquid crystalline host matrices. The encapsulation of peptides and proteins into such lyotropic liquid crystal host matrices has been shown to improve the stability and functionality of these molecules, an important aspect for biomedical applications (40, 41, 42).
In conclusion, the structure of a FRET biosensor based on the YFP-CFP FRET pair connected by a linker peptide for NE was studied in solution. The combination of SAXS with model-independent data analysis, DLS, and biochemical methods provided detailed information on the protein conformation in a native state in solution. The nanostructure of the intact biosensor, studied with synchrotron SAXS, indicates that the two proteins are preferably in parallel alignment, with a relative shift along the major axis. The flexibility in alignment of the subunits of the biosensor may also influence the FRET ratio and efficacy of the sensor. After reaction with NE, SAXS confirmed the decrease in biosensor size and shape deviations from ellipsoidal to more spherical in agreement with the shape expected from the individual proteins from the crystal structure. The modified fluorescence emission spectra of the biosensor before and after reaction confirmed its response to NE, in agreement with the changes in conformation of the fluorescent units from SAXS analysis.
The unique insights into protein conformation, folding state, size, and shape, together with the biochemical characterization of the sensor activity, allowed the correlation between nanostructure and function in the protein biosensor. Our results highlight SAXS combined with advanced data analysis as a powerful tool to characterize proteins in solution and investigate protein-protein interactions; it may also be valuable for the design of protein-based biosensors and the study of stimuli-responsive protein interactions, conformation, and morphology.
Author Contributions
Both authors contributed to the planning of experiments, interpretation of results, and writing of this article. G.F. handled expression of the biosensor. S.S. handled SAXS experiments and related data analysis.
Acknowledgments
The SAXS studies in this article were conducted at the Austrian SAXS beamline at the Elettra synchrotron station, Trieste, Italy. The authors thank Heinz Amenitsch for technical support and fruitful discussion at the beamline, and Luzia Wiesli and Thomas Ramsauer for technical assistance in overexpression and purification of the proteins.
Editor: Tommy Nylander.
Footnotes
Two figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30967-0.
Contributor Information
Greta Faccio, Email: greta.faccio@gmail.com.
Stefan Salentinig, Email: stefan.salentinig@empa.ch.
Supporting Material
References
- 1.Zadran S., Standley S., Baudry M. Fluorescence resonance energy transfer (FRET)-based biosensors: visualizing cellular dynamics and bioenergetics. Appl. Microbiol. Biotechnol. 2012;96:895–902. doi: 10.1007/s00253-012-4449-6. [DOI] [PubMed] [Google Scholar]
- 2.Ibraheem A., Campbell R.E. Designs and applications of fluorescent protein-based biosensors. Curr. Opin. Chem. Biol. 2010;14:30–36. doi: 10.1016/j.cbpa.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 3.Frommer W.B., Davidson M.W., Campbell R.E. Genetically encoded biosensors based on engineered fluorescent proteins. Chem. Soc. Rev. 2009;38:2833–2841. doi: 10.1039/b907749a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsien R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 5.Belousov V.V., Fradkov A.F., Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 6.Koay M.S., Janssen B.M.G., Merkx M. Tuning the metal binding site specificity of a fluorescent sensor protein: from copper to zinc and back. Dalton Trans. 2013;42:3230–3232. doi: 10.1039/c2dt32082g. [DOI] [PubMed] [Google Scholar]
- 7.Moussa R., Baierl A., Pohl M. An evaluation of genetically encoded FRET-based biosensors for quantitative metabolite analyses in vivo. J. Biotechnol. 2014;191:250–259. doi: 10.1016/j.jbiotec.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Piston D.W., Kremers G.J. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem. Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Zhang B. Design of FRET-based GFP probes for detection of protease inhibitors. Biochem. Biophys. Res. Commun. 2004;323:674–678. doi: 10.1016/j.bbrc.2004.08.143. [DOI] [PubMed] [Google Scholar]
- 10.Fehr M., Okumoto S., Frommer W.B. Development and use of fluorescent nanosensors for metabolite imaging in living cells. Biochem. Soc. Trans. 2005;33:287–290. doi: 10.1042/BST0330287. [DOI] [PubMed] [Google Scholar]
- 11.Vinkenborg J.L., Nicolson T.J., Merkx M. Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat. Methods. 2009;6:737–740. doi: 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demaurex N., Frieden M. Measurements of the free luminal ER Ca(2+) concentration with targeted “cameleon” fluorescent proteins. Cell Calcium. 2003;34:109–119. doi: 10.1016/s0143-4160(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 13.Jubany-Mari T., Alegre-Batlle L., Feldman L.J. Use of a redox-sensing GFP (c-roGFP1) for real-time monitoring of cytosol redox status in Arabidopsis thaliana water-stressed plants. FEBS Lett. 2010;584:889–897. doi: 10.1016/j.febslet.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Angres B., Steuer H., Schneckenburger H. A membrane-bound FRET-based caspase sensor for detection of apoptosis using fluorescence lifetime and total internal reflection microscopy. Cytometry A. 2009;75:420–427. doi: 10.1002/cyto.a.20698. [DOI] [PubMed] [Google Scholar]
- 15.Sekar R.B., Periasamy A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J. Cell Biol. 2003;160:629–633. doi: 10.1083/jcb.200210140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Roessel P., Brand A.H. Imaging into the future: visualizing gene expression and protein interactions with fluorescent proteins. Nat. Cell Biol. 2002;4:E15–E20. doi: 10.1038/ncb0102-e15. [DOI] [PubMed] [Google Scholar]
- 17.Schulenburg C., Faccio G., Richter M. A FRET-based biosensor for the detection of neutrophil elastase. Analyst (Lond.) 2016;141:1645–1648. doi: 10.1039/c5an01747e. [DOI] [PubMed] [Google Scholar]
- 18.O’Donoghue A.J., Jin Y., Hermiston T.W. Global substrate profiling of proteases in human neutrophil extracellular traps reveals consensus motif predominantly contributed by elastase. PLoS One. 2013;8:e75141. doi: 10.1371/journal.pone.0075141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nogales E. The development of cryo-EM into a mainstream structural biology technique. Nat. Methods. 2016;13:24–27. doi: 10.1038/nmeth.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yee A.A., Semesi A., Arrowsmith C.H. Screening proteins for NMR suitability. Methods Mol. Biol. 2014;1140:169–178. doi: 10.1007/978-1-4939-0354-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kikhney A.G., Svergun D.I. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015;589(19 Pt A):2570–2577. doi: 10.1016/j.febslet.2015.08.027. [DOI] [PubMed] [Google Scholar]
- 22.Mertens H.D., Piljić A., Svergun D.I. Conformational analysis of a genetically encoded FRET biosensor by SAXS. Biophys. J. 2012;102:2866–2875. doi: 10.1016/j.bpj.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amenitsch H., Rappolt M., Bernstorff S. First performance assessment of the small-angle X-ray scattering beamline at ELETTRA. J. Synchrotron Radiat. 1998;5:506–508. doi: 10.1107/S090904959800137X. [DOI] [PubMed] [Google Scholar]
- 24.Hammersley, A. P. 1998. ESRF Internal Report. ESRF98HA01T, FIT2D V9.129, http://www.esrf.eu/computing/scientific/FIT2D/FIT2D_REF/fit2d_r.html/.
- 25.Fritz G., Glatter O. Structure and interaction in dense colloidal systems: evaluation of scattering data by the generalized indirect fourier transformation method. J. Phys. Condens. Matter. 2006;18:2403–2419. [Google Scholar]
- 26.Glatter O. A new method for the evaluation of small-angle scattering data. J. Appl. Cryst. 1977;10:415–421. [Google Scholar]
- 27.Glatter O. The interpretation of real-space information from small-angle scattering experiments. J. Appl. Cryst. 1979;12:166–175. [Google Scholar]
- 28.Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svergun D.I., Petoukhov M.V., Koch M.H. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozin M.B., Svergun D.I. Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 2001;34:33–41. [Google Scholar]
- 31.Doniach S. Changes in biomolecular conformation seen by small angle X-ray scattering. Chem. Rev. 2001;101:1763–1778. doi: 10.1021/cr990071k. [DOI] [PubMed] [Google Scholar]
- 32.Bajar B.T., Wang E.S., Chu J. A guide to fluorescent protein FRET pairs. Sensors (Basel) 2016;16:1488. doi: 10.3390/s16091488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seegar T., Barton W. Imaging protein-protein interactions in vivo. J. Vis. Exp. 2010;44:2149. doi: 10.3791/2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hukasova E., Silva Cascales H., Lindqvist A. Monitoring kinase and phosphatase activities through the cell cycle by ratiometric FRET. J. Vis. Exp. 2012;59:e3410. doi: 10.3791/3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Millington M., Grindlay G.J., Magennis S.W. High-precision FLIM-FRET in fixed and living cells reveals heterogeneity in a simple CFP-YFP fusion protein. Biophys. Chem. 2007;127:155–164. doi: 10.1016/j.bpc.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Salentinig S., Phan S., Gilbert E.P. pH-responsive micelles based on caprylic acid. Langmuir. 2014;30:7296–7303. doi: 10.1021/la500835e. [DOI] [PubMed] [Google Scholar]
- 37.Mittelbach R., Porod G. Zur Röntgenkleinwinkelstreuung Verdünnter Systeme VII. Die Berechnung der Streukurven von Dreiachsigen Ellipsoiden. Acta Phys. Austriaca. 1962;15:122. [Google Scholar]
- 38.Receveur-Brechot V., Durand D. How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 2012;13:55–75. doi: 10.2174/138920312799277901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang F., Moss L.G., Phillips G.N., Jr. The molecular structure of green fluorescent protein. Nat. Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 40.Gontsarik M., Buhmann M.T., Salentinig S. Antimicrobial peptide-driven colloidal transformations in liquid-crystalline nanocarriers. J. Phys. Chem. Lett. 2016;7:3482–3486. doi: 10.1021/acs.jpclett.6b01622. [DOI] [PubMed] [Google Scholar]
- 41.Nylander T., Mattisson C., Hakansson B. A study of entrapped enzyme stability and substrate diffusion in a monoglyceride-based cubic liquid crystalline phase. Colloids Surf. A Physicochem. Eng. Asp. 1996;114:311–320. [Google Scholar]
- 42.Sun W., Vallooran J.J., Mezzenga R. Lyotropic liquid crystalline cubic phases as versatile host matrices for membrane-bound enzymes. J. Phys. Chem. Lett. 2016;7:1507–1512. doi: 10.1021/acs.jpclett.6b00416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.