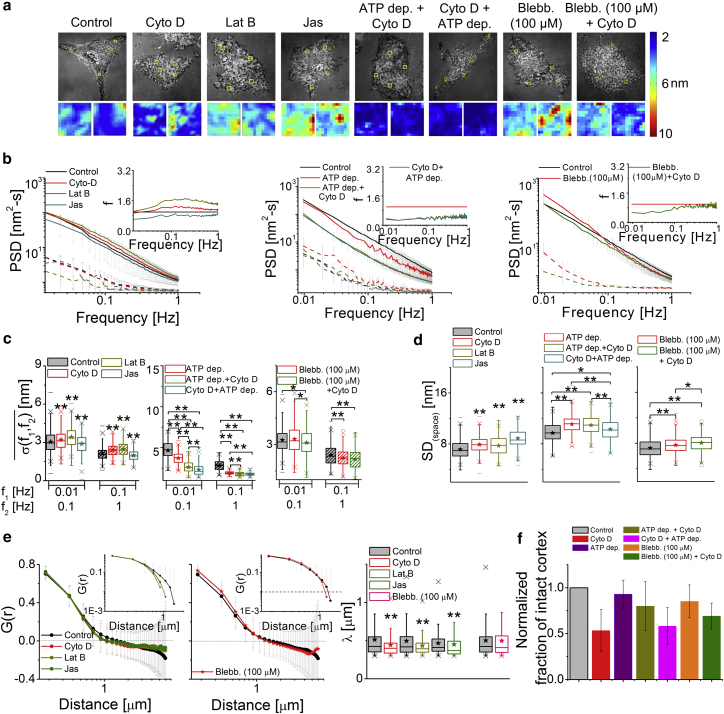
Figure 4.
Effect of the actomyosin cortex on membrane fluctuations. (a) Given here are IRM images (scale bars, 10 μm) and SD(time) maps (scale bars, 1 μm) of HeLa cells under mentioned conditions. (b) Shown here are PSDs for cells (solid lines) and their backgrounds (dashed lines), with the inset showing the value f for all conditions. (Left) Shown here are N = 30 cells each, ncontrol = 741 FBRs, nCyto D = 564 FBRs, nLat B = 426 FBRs, and nJas = 259 FBRs. (Middle) Shown here are N = 10 cells each, ncontrol = 141 FBRs, nATP depletion = 158 FBRs, nATP depletion + Cyto D = 254 FBRs, and nCyto D + ATP depletion = 162 FBRs. (Right) Shown here are N = 20 cells each, ncontrol = 402 FBRs, nblebbistatin = 333 FBRs, and nblebbistatin + Cyto D = 193 FBRs. (c and d) The parameters of temporal fluctuations and spatial undulations of cells are given under different conditions. ∗p < 0.05, ∗∗p < 0.001, one-way ANOVA. (e) Given here are averaged spatial ACFs (and their log-log plots, top inset) and correlations lengths (ncontrol = 1085 FBRs, nCyto D = 494 FBRs, N = 40 cells each; ncontrol = 884 FBRs, nLat B = 495 FBRs, N = 30 cells; ncontrol = 300 FBRs, nJas = 102 FBRs, N = 20 cells; and ncontrol = 811 FBRs, nblebbistatin = 729 FBRs, N = 30 cells). ∗p < 0.05, ∗∗p < 0.001, Mann-Whitney U test. (f) Given here is the normalized fraction of cortex clearance (N = 10 cells each). See also Figs. S8, S9, S10, S11, and S12; Tables S2 and S3. To see this figure in color, go online.