Figure 7.
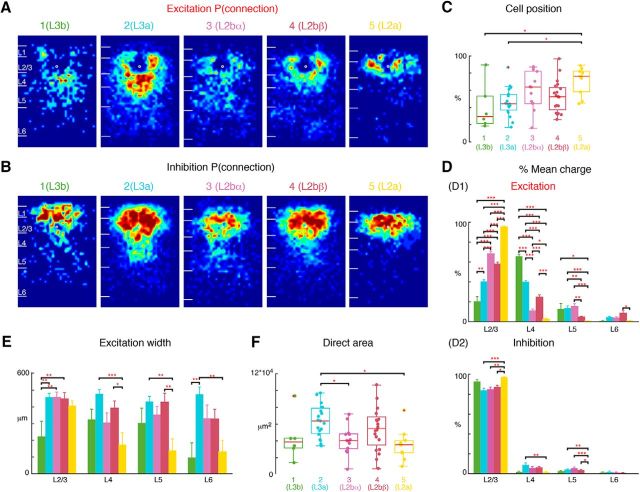
L2b further subdivides into two sublayers: L2bα, L2bβ. L2b cells in Figure 5 can be divided into additional two groups L2bα and L2bβ, resulting in five groups of L2/3 cells. The first two subgroups represent L3 (Group 1 = L3b, Group 2 = L3a; same as Fig. 6), whereas L2 consists of three subgroups (Group 3: L2bα, Group 4: L2bβ, Group 5: L2a; same as Fig. 5). A, Average maps (aligned to soma, white circle) of connection probability for excitatory connections among five groups of cells. Connection probability is encoded according to the pseudocolor scale. White horizontal lines indicate averaged laminar borders and are 100 μm long. B, Average maps (aligned to soma, white circle) of connection probability for inhibitory connections among five groups of cells. Connection probability is encoded according to the pseudocolor scale. White horizontal lines indicate averaged laminar borders and are 100 μm long. C, Boxplot of relative cell positions of Group 1 (green), Group 2 (light blue), Group 3 (purple), Group 4 (red), and Group 5 (yellow) cells within L2/3. The locations of L2a cells are close to the upper boundary of L2/3 and the locations of L3b and L3a cells are close to the lower boundary. The cell location in Groups 3 and 4 are in between. *p < 0.05, **p < 0.01. The p values from multiple-comparison tests are as follows: L3b vs L3a: p = 0.95; L3b vs L2bα: p = 0.21; L3b vs L2bβ: p = 0.63; L3b vs L2a: p = 0.02; L3a vs L2bα: p = 0.27; L3a vs L2bβ: p = 0.85; L3a vs L2a: p = 0.012; L2bα vs L2bβ: p = 0.78; L2bα vs L2a: p = 0.66; L2bβ vs L2a: p = 9.68 × 10−2. D, Layer-specific fractional excitatory (D1) and inhibitory (D2) charge of L3b (green), L3a (orange), L2bα (blue), L2bβ (light blue) and L2a (red) cells. Data are mean charge ± SEM. Comparing between each two groups the excitation originating from L2/3 or L4 all shows significance. The main input to L3b cells comes from L4, whereas the main input to L2a is within L2/3. L3a has most input both coming from L2/3 and L4. L2bα has input primarily coming from L2/3 and L5. The p values from Multi-comparison test are as follows: L2/3: L3b vs L3a: p = 2.34 × 10−5; L3b vs L2bα: p = 9.92 × 10−9; L3b vs L2bβ: p = 9.92 × 10−9; L3b vs L2a: p = 9.92 × 10−9; L3a vs L2bα: p = 9.92 × 10−9; L3a vs L2bβ: p = 9.79 × 10−8; L3a vs L2a: p = 9.92 × 10−9; L2bα vs L2bβ: p = 6.62 × 10−3; L2bα vs L2a: p = 1.06 × 10−8; L2bβ vs L2a: p = 9.92 × 10−9. L4: L3b vs L3a: p = 1.04 × 10−8; L3b vs L2bα: p = 9.92 × 10−9; L3b vs L2bβ: p = 9.92 × 10−9; L3b vs L2a: p = 9.92 × 10−9; L3a vs L2bα: p = 9.92 × 10−9; L3a vs L2bβ: p = 3.39 × 10−8; L3a vs L2a: p = 9.92 × 10−9; L2bα vs L2bβ: p = 4.39 × 10−6; L2bα vs L2a: p = 3.91 × 10−2; L2bβ vs L2a: p = 1.04 × 10−8. L5: L3b vs L3a: p = 1; L3b vs L2bα: p = 0.96; L3b vs L2bβ: p = 0.21; L3b vs L2a: p = 3.41 × 10−2; L3a vs L2bα: p = 0.97; L3a vs L2bβ: p = 7.83 × 10−3; L3a vs L2a: p = 7.83 × 10−4; L2bα vs L2bβ: p = 3.44 × 10−3; L2bα vs L2a: p = 3.62 × 10−4; L2bβ vs L2a: p = 0.66. L6: L3b vs L3a: p = 0.65; L3b vs L2bα: p = 0.82; L3b vs L2bβ: p = 5.2 × 10−2; L3b vs L2a: p = 1; L3a vs L2bα: p = 1; L3a vs L2bβ: p = 0.25; L3a vs L2a: p = 0.51; L2bα vs L2bβ: p = 0.21; L2bα vs L2a: p = 0.74; L2bβ vs L2a: p = 0.01. Inhibition also shows differences among the groups. L2a has most input from L2/3 and little originating from other layers compared with L3a, L2bα, and L2bβ; The p values from multiple-comparison tests of inhibition are as follows: L2/3: L3b vs L3a: p = 0.11; L3b vs L2bα: p = 0.25; L3b vs L2bβ: p = 0.53; L3b vs L2a: p = 0.81; L3a vs L2bα: p = 1; L3a vs L2bβ: p = 0.66; L3a vs L2a: p = 3.98 × 10−4; L2bα vs L2bβ: p = 0.93; L2bα vs L2a: p = 3.50 × 10−3; L2bβ vs L2a: p = 0.012. L4: L3b vs L3a: p = 6.43 × 10−2; L3b vs L2bα: p = 9.92 × 10−9; L3b vs L2bβ: p = 9.92 × 10−9; L3b vs L2a: p = 9.92 × 10−9; L3a vs L2bα: p = 0.61; L3a vs L2bβ: p = 0.58; L3a vs L2a: p = 8.77 × 10−3; L2bα vs L2bβ: p = 1; L2bα vs L2a: p = 0.35; L2bβ vs L2a: p = 0.17. L5: L3b vs L3a: p = 0.72; L3b vs L2bα: p = 0.28; L3b vs L2bβ: p = 0.92; L3b vs L2a: p = 0.58; L3a vs L2bα: p = 0.80; L3a vs L2bβ: p = 0.97; L3a vs L2a: p = 8.61 × 10−3; L2bα vs L2bβ: p = 0.44; L2bα vs L2a: p = 9.16 × 10−4; L2bβ vs L2a: p = 0.034. L6: L3b vs L3a: p = 0.93; L3b vs L2bα: p = 1; L3b vs L2bβ: p = 1; L3b vs L2a: p = 0.88; L3a vs L2bα: p = 0.86; L3a vs L2bβ: p = 0.57; L3a vs L2a: p = 0.20; L2bα vs L2bβ: p = 1; L2bα vs L2a: p = 0.76; L2bβ vs L2a: p = 0.87. E, Barplot of the distance of 80% of input to each L2/3 cell originating from L2/3, L4, L5, and L6. The p values from multiple-comparison test are as follows: L2/3: L3b vs L3a: p = 4.59 × 10−3; L3b vs L2bα: p = 9.62 × 10−3; L3b vs L2bβ: p = 7.53 × 10−3; L3b vs L2a: p = 8.32 × 10−2; L3a vs L2bα: p = 1; L3a vs L2bβ: p = 1; L3a vs L2a: p = 0.88; L2bα vs L2bβ: p = 1; L2bα vs L2a: p = 0.92; L2bβ vs L2a: p = 0.95. L4: L3b vs L3a: p = 0.38; L3b vs L2bα: p = 1; L3b vs L2bβ: p = 0.93; L3b vs L2a: p = 0.48; L3a vs L2bα: p = 6.24 × 10−2; L3a vs L2bβ: p = 0.56; L3a vs L2a: p = 5.06 × 10−4; L2bα vs L2bβ: p = 0.63; L2bα vs L2a: p = 0.44; L2bβ vs L2a: p = 0.022. L5: L3b vs L3a: p = 0.63; L3b vs L2bα: p = 0.99; L3b vs L2bβ: p = 0.67; L3b vs L2a: p = 0.49; L3a vs L2bα: p = 0.77; L3a vs L2bβ: p = 1; L3a vs L2a: p = 2.91 × 10−3; L2bα vs L2bβ: p = 0.81; L2bα vs L2a: p = 0.95; L2bβ vs L2a: p = 3.43 × 10−3. L6: L3b vs L3a: p = 9.89 × 10−3; L3b vs L2bα: p = 0.29; L3b vs L2bβ: p = 0.24; L3b vs L2a: p = 1; L3a vs L2bα: p = 0.43; L3a vs L2bβ: p = 0.30; L3a vs L2a: p = 3.90 × 10−3; L2bα vs L2bβ: p = 1; L2bα vs L2a: p = 0.29; L2bβ vs L2a: p = 0.22. F, Boxplot of the direct activation area of the cell groups. L3a cells have bigger direct activation area than group L2bα and L2a cells (L3a vs L2bα: p = 0.037; L3a vs L2a: p = 0.011). There is no difference between the other groups (L3b vs L3a: p = 0.27; L3b vs L2bα: p = 0.99; L3b vs L2bβ: p = 0.80; L3b vs L2a: p = 0.95; L3a vs L2bβ: p = 0.67; L2bα vs L2bβ: p = 0.41; L2bα vs L2a: p = 0.97). Triple asterisk indicates a higher significance level.