ABSTRACT
The Arabidopsis PEN3 ABC transporter accumulates at sites of pathogen detection, where it is involved in defense against a number of pathogens. Perception of PAMPs by pattern recognition receptors initiates recruitment of PEN3 and also leads to PEN3 phosphorylation at multiple amino acid residues. Whether PAMP-induced phosphorylation of PEN3 is important for its defense function or focal recruitment has not been addressed. In this study, we evaluated the role of PEN3 phosphorylation in modulating the localization and defense function of the transporter. We report that PEN3 phosphorylation is critical for its function in defense, but dispensable for recruitment to powdery mildew penetration sites. These results indicate that PAMP-induced phosphorylation is likely to regulate the transport activity of PEN3.
KEYWORDS: Defense response, powdery mildew, pattern-triggered immunity, pattern-recognition receptor, papilla, signal transduction
The plant antimicrobial immune system is activated upon detection of potentially pathogenic microorganisms mediated primarily by two major classes of immune receptors, pattern recognition receptors (PRRs) that perceive conserved elicitor molecules characteristic of microorganisms, termed pathogen- or microbe-associated molecular patterns (PAMPs/MAMPs), or nucleotide-binding leucine-rich repeat (NB-LRR) type resistance (R) proteins that detect pathogen virulence factors or their activities.7,11,13,14 Stimulation of PRRs by PAMP perception results in the activation of a number of characteristic defense outputs, including local reinforcement of the cell wall through construction of papillae, a process that involves deposition of callose, accumulation of antimicrobial metabolites, and recruitment of defense-associated proteins.21-23 The Arabidopsis PENETRATION3 (PEN3; synonyms ABCG36 and PDR8) ATP-binding cassette (ABC) transporter is required for full resistance to cell wall penetration and subsequent haustorium formation by the non-host barley powdery mildew pathogen Blumeria graminis f. sp. hordei (Bgh) and contributes to resistance to numerous other fungal and oomycete pathogens.8,9,11,12,18 PEN3 is recruited to sites of papilla deposition upon perception of PAMPs and is required for full pattern-triggered immunity against the bacterial pathogen Pseudomonas syringae pv. tomato DC3000, indicating that the pathway involving the PEN3 transporter participates in the pattern-triggered immune response.22,24 In accordance with these findings, phosphoproteomics studies identified amino acid residues of the PEN3 transporter that were differentially phosphorylated in response to perception of the PAMPs flg22, an elicitor active peptide derived from bacterial flagellin, oligo-galacturonide (OG) fragments derived from the cell wall polymer pectin, or fungal xylanase.4,10,15 However, whether PAMP-induced phosphorylation of PEN3 is required for the defense function of the transporter has not been addressed. In this study, we generated mutant PEN3 variants at proteomically supported phosphorylation sites and evaluated their ability to restore penetration defense in the pen3 mutant and to be recruited to powdery mildew penetration sites.
Several phosphoproteomic studies of Arabidopsis PM proteins have identified phosphorylation sites in the PEN3 transporter.4,10,15,16 An evaluation of PM phosphoproteins enriched from a mixture of control- and flg22-treated suspension culture cells unambiguously identified phosphorylation events at serine residues S37, S40, S45, S841, and S844 as well as threonine residue T43.14 Additionally, serine residue S38 was identified as a putative phosphorylated residue that could not be unambiguously identified. Subsequent quantitative studies revealed that serine residues S40 and S45 are differentially phosphorylated in response to perception of flg22 or fungal xylanase and identified an additional serine residue, S825, that is marginally differentially phosphorylated (significant at P = 0.1)in response to flg22 treatment.4,15 Additionally, serines S37 and S38 could not be unambiguously ruled out as potential sites of differential phosphorylation in response to flg22.15 Similarly, quantitative analysis of phosphorylation events in response to perception of OGs by Arabidopsis seedlings identified differential PEN3 phosphorylation at a single ambiguous residue, either serine S37 or S40.10
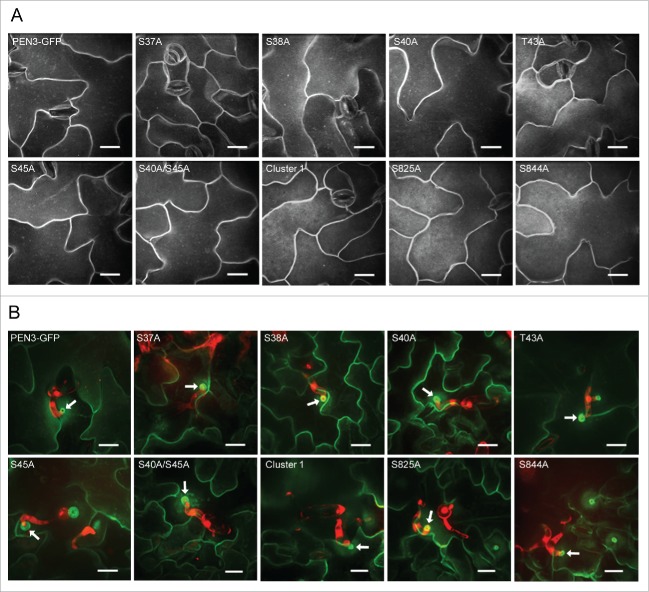
We hypothesized that PAMP-induced phosphorylation of PEN3 may contribute to the activation of the PEN3 defense function or to its recruitment to sites of interaction with invading pathogens. To evaluate the potential roles of specific PEN3 phosphorylated residues, we used site-directed mutagenesis to generate a series of alanine substitution mutants of an existing PEN3 promoter-PEN3-GFP construct,18 replacing each definitively or putatively phosphorylated residue with alanine to mimic the de-phosphorylated state. Additionally, we created three polymutants. PEN3 phosphorylation sites are not evenly distributed throughout the protein, but fall into two distinct clusters; an N-terminal cluster consisting of serine residues S37, S38, S40, and S45 as well as threonine T43, and a central cluster consisting of serine residues S825, S841, and S844 (Fig. 1A). We created polymutants containing alanine substitutions at each residue in the two distinct clusters. We refer to these polymutants as “Cluster 1” (5 N-terminal residues) and “Cluster 2” (3 central serines). We also created a double mutant containing alanine substitutions at the PAMP-responsive differentially phosphorylated residues, S40 and S45. We transformed each mutant construct into the pen3-3 mutant (SALK_110926;1,18) and assessed the ability of each mutant PEN3 variant to restore penetration resistance in response to Bgh at 48 hours post-inoculation (hpi) by creating transgenic plants and evaluating fungal penetration success as previously described.22 The PEN3 variants S37A, S38A, T43A, S825A, and S844A all were able to restore full penetration resistance against Bgh to the pen3-3 mutant (Fig. 1B), indicating that phosphorylation at these residues is not required for the defense function of PEN3. The PEN3 S40A and S45A variants failed to restore penetration resistance (Fig. 1B), suggesting that phosphorylation at these residues is important for the defense activity of the transporter. Consistent with a requirement for phosphorylation at serines S40 and S45, the S40A/S45A double mutant and Cluster 1 polymutant also failed to restore penetration resistance (Fig. 1B). We were unable to recover any S841A or Cluster 2 polymutant transformants exhibiting detectable levels of GFP fluorescence, suggesting that phosphorylation at serine S841 may contribute to protein stability. All other alanine substitution PEN3 variants exhibited GFP fluorescence and localization to the PM that was indistinguishable from wild-type PEN3-GFP (Fig. 2A).
Figure 1.
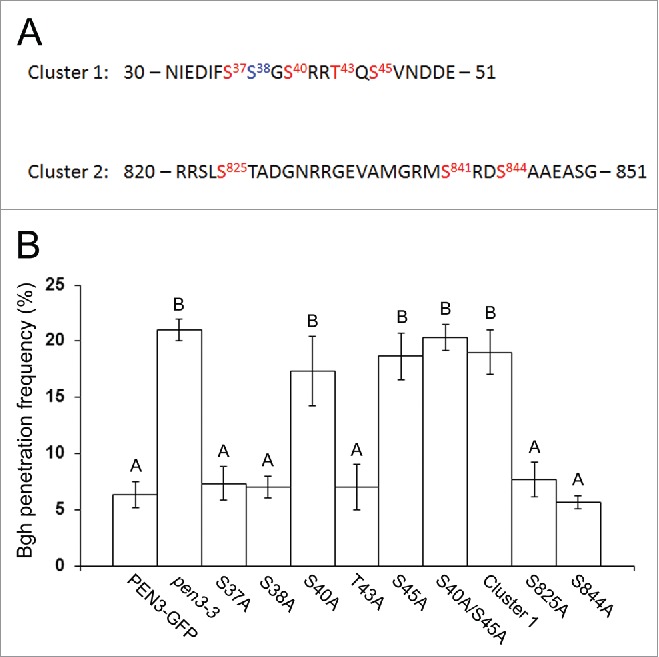
Complementation of pen3-3 by PEN3 phosphorylation site mutants. (A) PEN3 amino acid sequences in the regions encompassing phosphorylation sites at Cluster 1 (upper panel) and Cluster 2 (lower panel). Proteomically supported phosphorylation sites are colored red. Putative sites that lack unambiguous support are colored blue. (B) Bgh penetration frequencies on the wild-type PEN3-GFP line, pen3-3, and plants expressing phosphorylation site alanine substitution PEN3-GFP variants are expressed as means of the percentages of haustoria observed relative to total penetration attempts. Penetration frequencies were determined by scoring for presence or absence of haustoria at 100 infection sites per leaf for 3 leaves per plant line 48 hpi with Bgh. Error bars represent SD (n = 3). Means indicated by the same letter are not significantly different according to Tukey's post-hoc test (P < 0.01). The experiment was repeated 3 times with similar results.
Figure 2.
Localization of phosphorylation site alanine substitution variants of PEN3-GFP. (A) Confocal micrographs illustrating the expression and steady-state localization of PEN3-GFP alanine substitution variants in leaf epidermal cells. All variants exhibit PM localization similar to wild-type PEN3-GFP. (B) Confocal micrographs illustrating the localization of PEN3-GFP alanine substation variants in leaf epidermal cells at 24 hpi with Bgh. PEN3-GFP and variants appear green, propidium iodide-stained fungal structures appear red. Note that all alanine substitution variants exhibit focal accumulation at powdery mildew penetration sites similar to wild-type PEN3-GFP. At least 3 independent transgenic lines were evaluated for each alanine substitution variant with similar results. All panels are z-projected confocal images. Arrows indicate representative Bgh penetration sites. Scale bars = 20 μm.
PAMP-induced phosphorylation of PEN3 at serine residues S40 and S45 could potentially modulate the defense function of the transporter by activating or altering the transport activity of PEN3 or, alternatively, by affecting recruitment of PEN3 to sites of attempted Bgh penetration. To determine if disruption of PAMP-induced phosphorylation in our alanine substitution variants altered the ability of PEN3 to accumulate at sites of fungal penetration, we monitored the localization of each GFP-tagged PEN3 variant 24 h after inoculation with Bgh using confocal microscopy as previously described.22 We found that all of the detectable PEN3 alanine substitution variants retained the ability to accumulate at sites of attempted Bgh penetration (Fig. 2B). These results suggest that PAMP-induced phosphorylation likely contributes to the activation or alteration of PEN3 transport activity and is not required for recruitment of PEN3 to sites of attempted fungal penetration.
Phosphorylation of ABC transporters by protein kinases is well known to play a role in regulation of both transport activity and protein stability and turnover.2,19 Here, we identified two PEN3 phosphorylation sites, serines S40 and S45, required for the defense function of the transporter that are likely involved in regulating transport activity. In many instances where phosphorylation has been observed to regulate ABC transporter activity, relevant phosphorylation sites have been located within a regulatory (R) or R-like domain present in the cytosolic loop between the first nucleotide binding domain and the second transmembrane domain of the protein.19 Thus, our finding of relevant regulatory phosphorylation sites N-terminal to the first nucleotide binding domain within PEN3 contrasts with other previous observations that are primarily from humans or yeast. Nonetheless, the PAMP-responsive PEN3 phosphorylation sites are near the first nucleotide binding domain, which is consistent with numerous reports of regulatory phosphorylation events at sites within or adjacent to nucleotide binding domains of other ABC transporters.19 Additionally, a third PEN3 phosphorylation site, serine S841, appears to affect protein stability. A synthetic peptide encompassing serines S37, S38, S40, S45, and threonine T43 was identified as an in vitro substrate of the Arabidopsis calcium-dependent protein kinase CPK10.6 Further work will be required to determine if CPK10 plays a role in the regulation of PEN3 activity upon PAMP perception.
PEN3 is thought to contribute to antimicrobial defense by potentially transporting one or more molecules derived from metabolism of indole glucosinolates,3,5 however, the specific defense-relevant molecule(s) transported by PEN3 remain unknown. Additionally, PEN3 affects sensitivity of Arabidopsis to the auxin analog 2,4-DB and has been proposed to act redundantly with ABCG37 to transport the auxin precursor indole-3-butyric acid.17,20 It will be interesting to determine whether phosphorylation at S40 and S45 also regulates transport of auxin-related molecules by PEN3 or if other phosphorylation events modulate PEN3 function in growth and development.
Disclosure of potential conflicts of interest
The authors report no conflict of interest.
Funding
This work was supported by National Science Foundation Grants 0519898 and 0929226; by National Institutes of Health Postdoctoral Fellowship Grant F32-GM-0834393; and by the USDA Agricultural Research Service CRIS Project 3060-21220-031-00D. USDA is an equal opportunity provider and employer.
References
- 1.Alonso JM, Stepanova AN, Leissa TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al.. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–7. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 2.Aryal B, Laurent C, Geisler M. Learning from each other: ABC transporter regulation by protein phosphorylation in plant and mammalian systems. Biochem Soc Trans. 2016;44:663–73. doi: 10.1042/BST20150128_2. [DOI] [PubMed] [Google Scholar]
- 3.Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al.. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–6. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- 4.Benschop JJ, Mohammed S, O'Flaherty M, Heck AJ, Menke FL. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics. 2007;6:1198–214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curren A, Chang IF, Chang CL, Garg S, Miguel RM, Barron YD, Li Y, Romanowsky S, Cushman JC, Gribskov M, et al.. Calcium-dependent protein kinases from Arabidopsis show substrate specificity differences in an analysis of 103 substrates. Front Plant Sci. 2011;2:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodds PN, Rathjen JP. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–48. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 8.Egusa M, Miwa T, Kaminaka H, Takano Y, Kodama M. Nonhost resistance of Arabidopsis thaliana against Alternaria alternata involves both pre- and postinvasive defenses but is collapsed by AAL-toxin in the absence of LOH2. Phytopathology. 2013;103:733–40. doi: 10.1094/PHYTO-08-12-0201-R. [DOI] [PubMed] [Google Scholar]
- 9.Hiruma K, Onozawa-Komori M, Takahashi F, Asakura M, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y. Entry mode-dependent function of an indole glucosinolate pathway in Arabidopsis for nonhost resistance against anthracnose pathogens. Plant Cell. 2010;22:2429–43. doi: 10.1105/tpc.110.074344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohorn BD, Hoon D, Minkoff BB, Sussman MR, Kohorn SL. Rapid oligo-galacturonide induced changes in protein phosphorylation in Arabidopsis. Mol Cell Protemics. 2016;15:1351–9. doi: 10.1074/mcp.M115.055368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–29. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 12.Loehrer M, Langenbach C, Goellner K, Conrath U, Schaffrath U. Characterization of nonhost resistance of Arabidopsis to the Asian soybean rust. Mol Plant Microbe Interact. 2008;21:1421–31. doi: 10.1094/MPMI-21-11-1421. [DOI] [PubMed] [Google Scholar]
- 13.Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: So far and yet so close. Nat Immunol. 2011;12:817–26. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- 14.Monaghan J, Zipfel C. Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol. 2012;15:349–57. doi: 10.1016/j.pbi.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Nühse TS, Bottrill AR, Jones AM, Peck SC. Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 2007;51:931–40. doi: 10.1111/j.1365-313X.2007.03192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nühse TS, Stensballe A, Jensen ON, Peck SC. Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell. 2004;16:2394–405. doi: 10.1105/tpc.104.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruzicka K, Strader LC, Bailly A, Yang H, Blakeslee J, Langowski L, Nejedlá E, Fujita H, Itoh H, Syono K, et al.. Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc Natl Acad Sci USA. 2010;107:10749–53. doi: 10.1073/pnas.1005878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18:731–46. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stolarczyk EI, Reiling CJ, Paumi CM. Regulation of ABC transporter function via phosphorylation by protein kinases. Curr Pharm Biotechnol. 2011;12:621–35. doi: 10.2174/138920111795164075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strader LC, Bartel B. The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell. 2009;21:1992–2007. doi: 10.1105/tpc.109.065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Underwood W. The plant cell wall: a dynamic barrier against pathogen invasion. Front Plant Sci. 2012;3:85. doi: 10.3389/fpls.2012.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Underwood W, Somerville SC. Perception of conserved pathogen elicitors at the plasma membrane leads to relocalization of the Arabidopsis PEN3 transporter. Proc Natl Acad Sci USA. 2013;110:12492–97. doi: 10.1073/pnas.1218701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voigt CA. Callose-mediated resistance to pathogenic intruders in plant defense-related papillae. Front Plant Sci. 2014;5:168. doi: 10.3389/fpls.2014.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xin XF, Nomura K, Underwood W, He SY. Induction and suppression of PEN3 focal accumulation during Pseudomonas syringae pv. tomato DC3000 infection of Arabidopsis. Mol Plant Microbe Interact. 2013;26:861–67. doi: 10.1094/MPMI-11-12-0262-R. [DOI] [PubMed] [Google Scholar]