FIGURE 1.
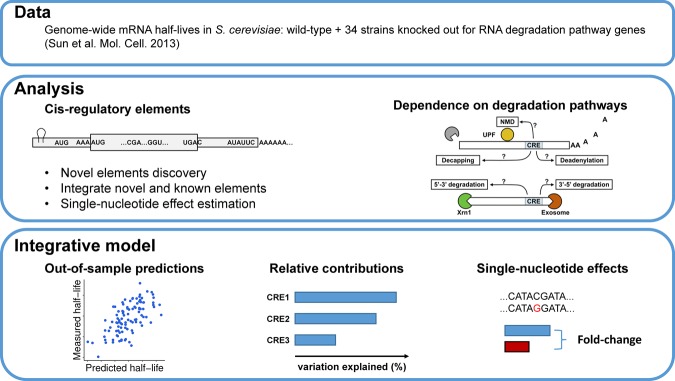
Study overview. The goal of this study is to discover and integrate cis-regulatory mRNA elements affecting mRNA stability and assess their dependence on mRNA degradation pathways. (Data) We obtained S. cerevisiae genome-wide half-life data from wild-type (WT) as well as from 34 knockout strains from Sun et al. (2013). Each of the knockout strains has one gene closely related to mRNA degradation pathways knocked out. (Analysis) We systematically searched for novel sequence features associating with half-life from 5′ UTR, start codon context, CDS, stop codon context, and 3′ UTR. Effects of previously reported cis-regulatory elements were also assessed. Moreover, we assessed the dependencies of different sequence features on degradation pathways by analyzing their effects on the knockout strains. (Integrative model) We built a statistical model to predict genome-wide half-life solely from mRNA sequence. This allowed the quantification of the relative contributions of the sequence features to the overall variation across genes and assessing the sensitivity of mRNA stability with respect to single-nucleotide variants.