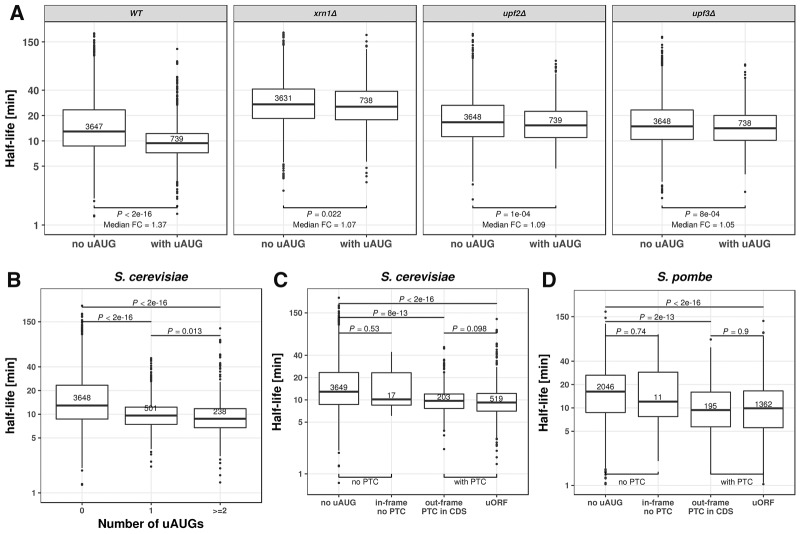
FIGURE 2.
Upstream AUG codons (uAUG) destabilize mRNA. (A) Distribution of mRNA half-lives for mRNAs without uAUG (left) and with at least one uAUG (right). From left to right: wild type, XRN1, UPF2, and UPF3 knockout S. cerevisiae strains. Median fold-change (Median FC) calculated by dividing the median of the group without uAUG with the group with uAUG. A complete view of the effect of uAUG across different knockouts is provided in Supplemental Figure S6. (B) Distribution of mRNA half-lives for mRNAs with zero (left), one (middle), or more (right) uAUGs in S. cerevisiae. (C) Distribution of mRNA half-lives for S. cerevisiae mRNAs with, from left to right: no uAUG, with one in-frame uAUG but no cognate premature termination codon, with one out-of-frame uAUG and one cognate premature termination codon in the CDS, and with one uAUG and one cognate stop codon in the 5′ UTR (uORF). (D) Same as in C for S. pombe mRNAs. All P-values were calculated with Wilcoxon rank-sum test. Numbers in the boxes indicate number of members in the corresponding group. Boxes represent quartiles, whiskers extend to the highest or lowest value within 1.5 times the interquartile range, and horizontal bars in the boxes represent medians. Data points falling further than 1.5-fold the interquartile distance are considered outliers and are shown as dots.