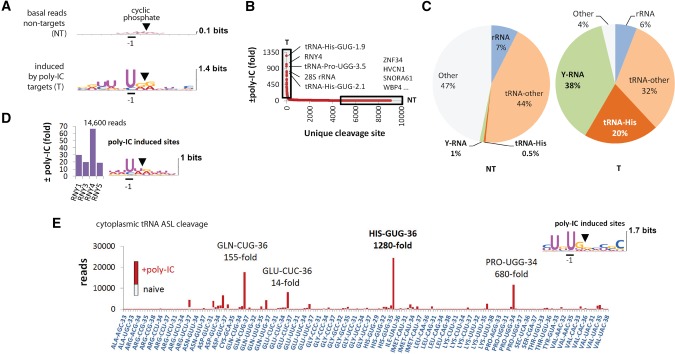
FIGURE 1.
RtcB RNA-seq analysis of RNA cleavage in poly(IC)-stimulated HeLa cells. (A) Sequence consensus at cleavage sites with 2′,3′-cyclic phosphates before and after poly(IC) treatment. Nontargets (NT) and targets (T) are defined in B. (B) Cleavage site distribution according to fold-induction by poly(IC). Boxed regions mark RNase L-sensitive (T) and resistant (NT) RNAs. (C) Composition of the NT and T groups by main RNA types. The group “other” contains primarily U6 small nucleolar RNA as well as mRNAs, micro-RNAs, and small ncRNAs (Supplemental Dataset S1). (D) Up-regulation of reads for each Y-RNA by poly(IC). Y-RNAs are cleaved at UN^N consensus sites. (E) ASL cleavage sites in observed fragments of cytosolic tRNAs. The stacked bars show basal (gray) and poly(IC)-induced (red) reads for each site.