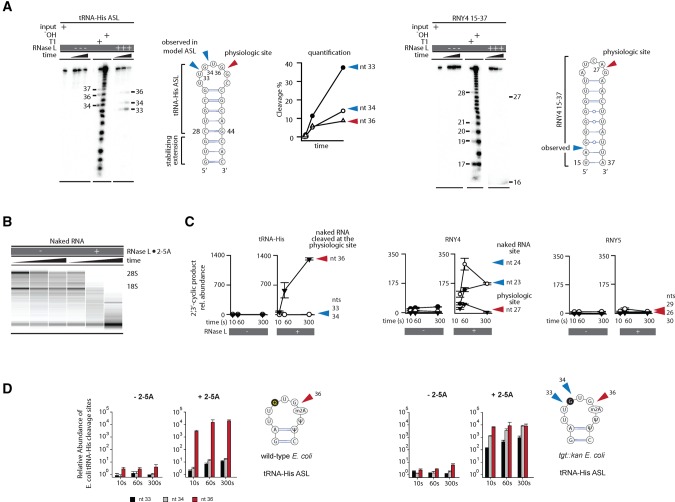
FIGURE 3.
Cleavage of naked human RNA points to distinct specificity determinants for Y-RNAs and tRNAs. (A) Cleavage of model stem–loops from tRNA-His and RNY4 analyzed by polyacrylamide gel electrophoresis (PAGE). Neither model substrate is cleaved preferentially at the physiologic site. (B) RNA chip analysis for cleavage of protein-free total RNA by RNase L. The nonspecific decay observed in this experiment contrasts the site-specific cleavage in cells (Supplemental Fig. S1B). (C) RtcB qPCR analysis to measure cleavage of tRNA-His and RNY4 at physiologic and nonphysiologic sites in naked RNA. Error bars show SE from two qPCR replicates. (D) Cleavage of tRNA-His purified from WT or mutant E.coli lacking a queuosine biosynthesis gene tgt was measured by RtcB qPCR. The queuosine position is shaded gray. Error bars show SE from two biological replicates.