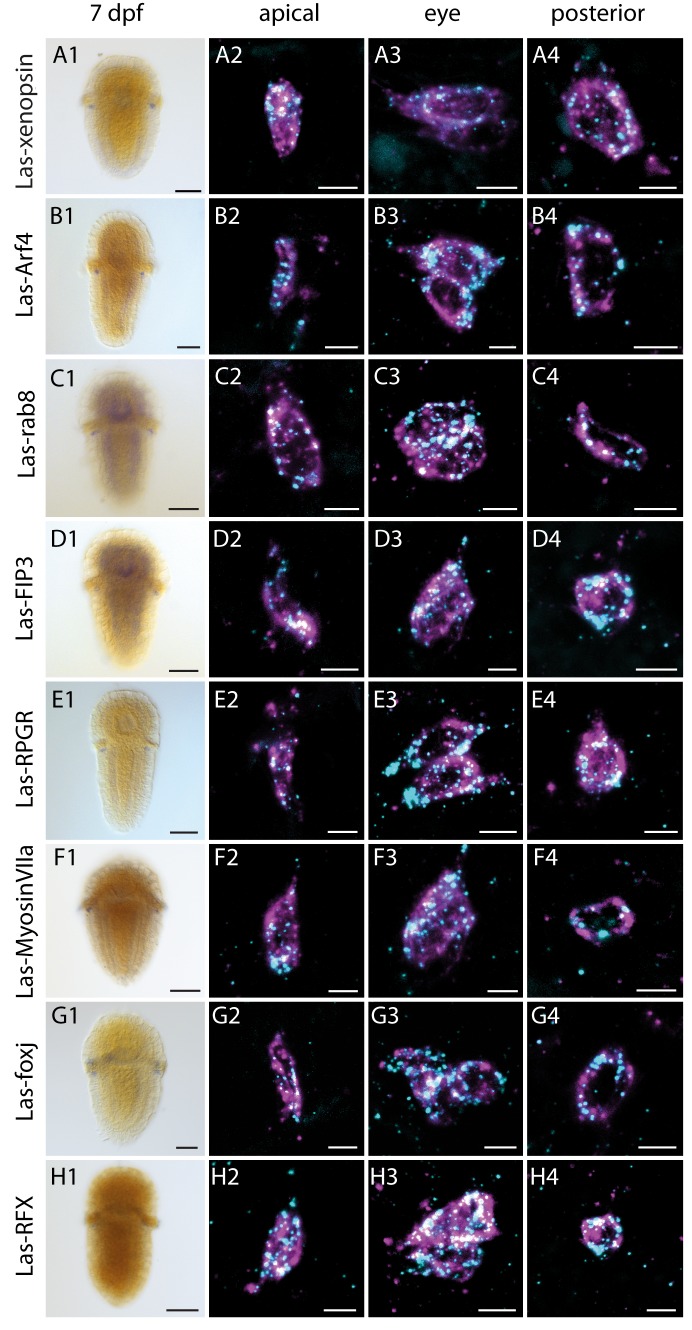
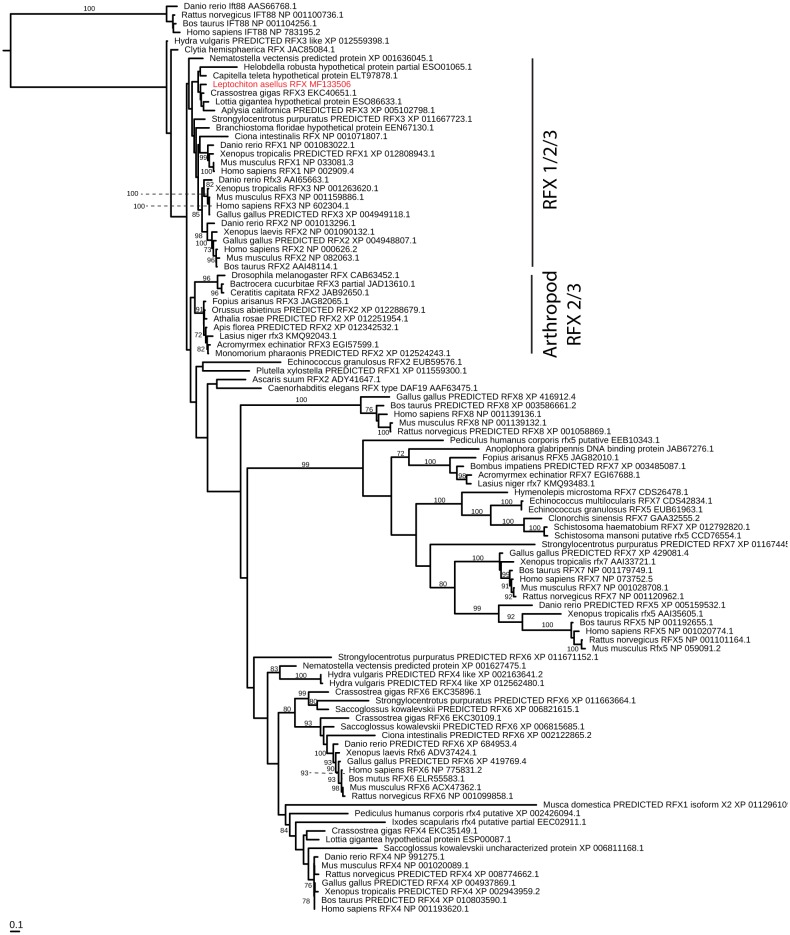
Figure 4. Expression of Las-xenopsin as well as genes involved in ciliary opsin transport and development of cilia.
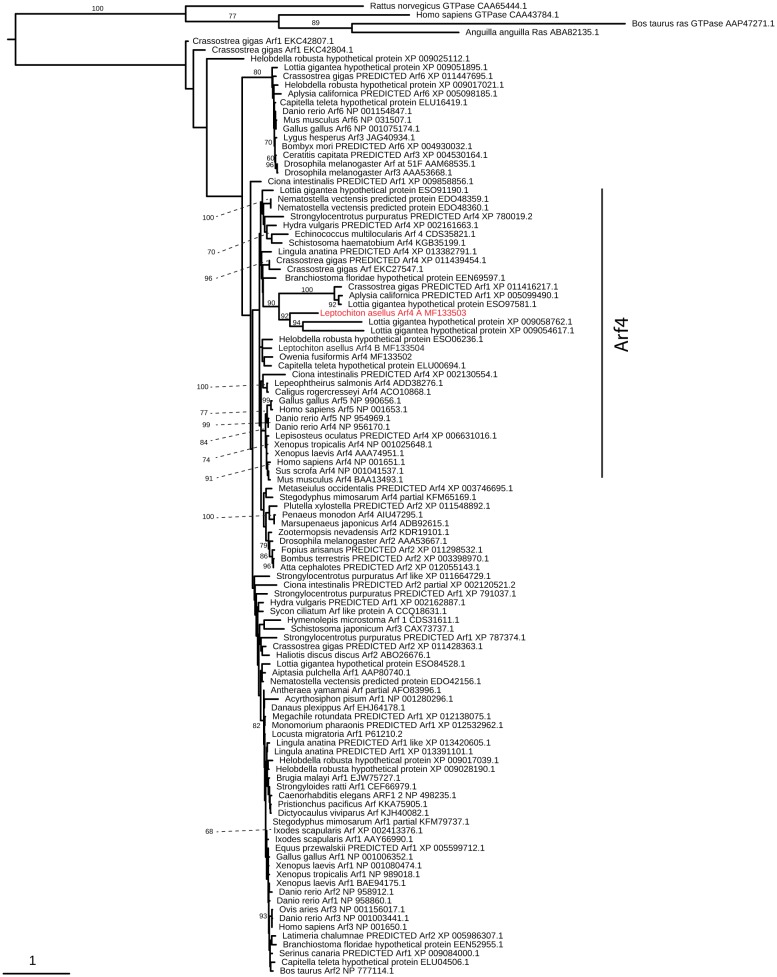
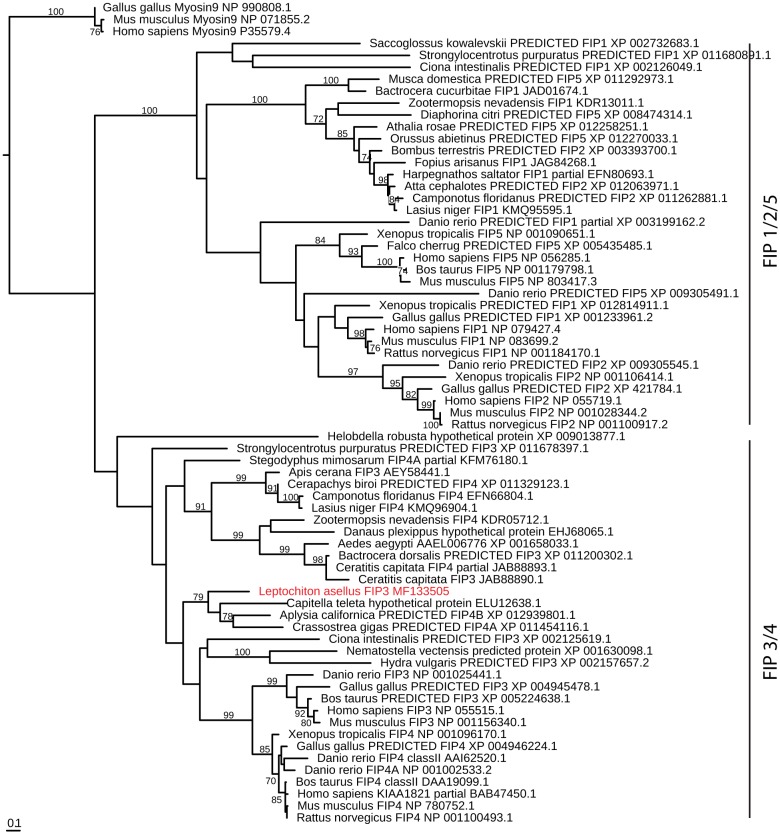
Column 1: single labeling of gene X. Column 2–4: double labeling of gene X (cyan) and Las-r-opsin (magenta) in the anterior, posttrochal eye and posterior regions. Las-xenopsin (a1–a4), Las-Arf4 (b1–b4), Las-rab8 (c1–c4), Las-FIP3 (d1-4), Las-RPGR (e1-4), Las-Myosin VIIa (f1-4), Las-foxj1 (g1-4) and Las-RFX (h1-4) are coexpressed with Las-r-opsin in anterior, eye and posterior PRCs. Additionally, Las-rab8 (c1) and Las-FIP3 (d1) show a broader expression in the nervous system of 7 dpf larvae and Las-foxj1 shows expression also in the apical area and prototroch cells of young larvae (Figure 4—figure supplement 1). See Figure 4—figure supplements 1–3 for gene trees of Arf4, FIP and RFX. (Scalebars: 100 μm in column 1; 5 μm in columns 2–4).
Figure 4—figure supplement 1. Las-foxj1 expression in a young larva (48 hpf).
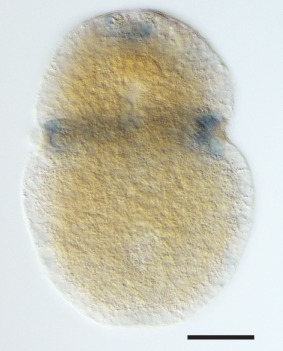
Figure 4—figure supplement 2. Alignment of C-termimal ends of the opsins involved in this study.