Abstract
The National Surgical Quality Improvement Project (NSQIP) is widely recognized as “the best in the nation” surgical quality improvement resource in the United States. In particular, it rigorously defines postoperative morbidity outcomes, including surgical adverse events occurring within 30 days of surgery. Due to its manual yet expensive construction process, the NSQIP registry is of exceptionally high quality, but its high cost remains a significant bottleneck to NSQIP’s wider dissemination. In this work, we propose an automated surgical adverse events detection tool, aimed at accelerating the process of extracting postoperative outcomes from medical charts. As a prototype system, we combined local EHR data with the NSQIP gold standard outcomes and developed machine learned models to retrospectively detect Surgical Site Infections (SSI), a particular family of adverse events that NSQIP extracts. The built models have high specificity (from 0.788 to 0.988) as well as very high negative predictive values (>0.98), reliably eliminating the vast majority of patients without SSI, thereby significantly reducing the NSQIP extractors’ burden.
Keywords: Electronic Health Records, National Surgical Quality Improvement Project, Supervised Learning, Surgical Site Infection
Introduction
The American College of Surgeons (ACS) National Surgical Quality Improvement Project (NSQIP) is widely recognized as “the best in the nation” surgical quality improvement resource in the United States [1]. NSQIP helps member hospitals to track outcomes associated with surgical patients, by collecting data on over 150 variables, including preoperative characteristics, intraoperative factors, and postoperative morbidity occurrences. In particular, postoperative morbidity outcomes are rigorously defined surgical adverse events occurring within 30 days of surgery, such as surgical site infection (SSI), urinary tract infection (UTI), and acute renal failure (ARF). NSQIP uses collected data elements to calculate relative performance regarding postoperative morbidity and mortality and to compare each member hospital’s performance with benchmarking, which is risk stratified, including providing an observed to expected (O/E ratio) for every surgical adverse event [2]. With this feedback, member hospitals are able to focus attention and resources to areas of opportunity for improving the care of patients, which may also result in achieving reduced length of stay and readmission rates [3].
Unfortunately, less than 20% of hospitals in the United States currently participate in NSQIP, in large part due to its associated costs to implement. In addition to the participation fee, hospitals must employ a formally trained surgical clinical reviewer (SCR). An SCR ensures the reliability of clinical data abstraction, selects operation cases following NSQIP inclusion criteria, manually reviews and extracts data elements, and documents surgical postoperative occurrence outcomes. This manual yet expensive approach leads to high-quality clinical data, but the associated cost remains a significant bottleneck to NSQIP’s wider dissemination.
An SSI is an infection occurring after surgery in the part of the body where surgery took place. While most surgical patients do not experience an SSI [4], SSIs are very expensive and morbid. According to the depth and severity of infection, SSIs are categorized into superficial, deep, and organ/space. Definitions for SSIs have been standardized by the Centers for Disease Control and Prevention (CDC) and are used by NSQIP SCR to identify and document each SSI category [5–6].
Previous work has explored risk factors associated with SSI, but few studies have focused on the detection of SSI. Most papers examining detection have relied heavily on administrative data or claims databases (such as age, gender, principal diagnosis, and billing information about medications and procedures) [7–8]. Since EHR data contains more detailed and richer clinical data (e.g. vital signs, lab results, and social history), compared with claims data it would provide additional significant indicators and signals to SSI and thus enhance the detection performance. In addition, most studies are procedure-specific, only processing SSIs following certain types of operation, such as hip and knee arthroplasty [9–10], instead of the current approach which is broadly inclusive of different types of surgery. To help reduce the labor and cost in reviewing patient records for postoperative surgical occurrences, we hypothesized that we could leverage both electronic health record (EHR) data and historic NSQIP registry data to develop and validate an automated approach with supervised machine learning algorithms to detect NSQIP occurrence outcomes. In particular, we focused on the postoperative SSI occurrences to develop a classifier of three SSI categories (superficial, deep, and organ/space) and the total SSI, and to reduce the SCR’s burden by eliminating the vast majority of patients associated with surgeries that did not result in SSI.
Materials and Methods
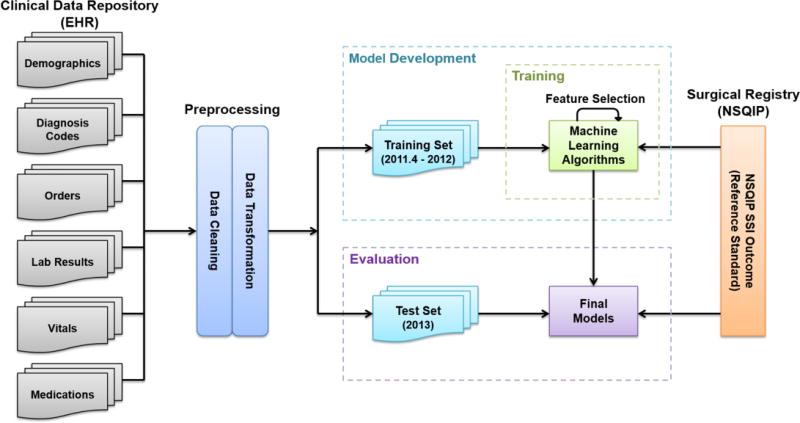
Our overall methodological approach for this study included four steps as outlined in Figure 1: (1) identification of the patient cohort and associated patient EHR data, (2) data preprocessing, (3) iterative supervised learning model development, and (4) evaluation of the final models using gold standard outcome data from the NSQIP registry. Institutional review board approval was obtained and informed consent waived for this minimal risk study.
Figure 1.
Overview of Materials and Methods
Data Collection and Patient Cohort Identification
The University of Minnesota Academic Health Center Information Exchange platform includes access to the clinical data repository (CDR) which contains University of Minnesota Medical Center (UMMC) clinical data. UMMC has been a member of NSQIP since 2007 and has used the inpatient instance of Epic since April 2011. CDR and NSQIP registry, two different data sources, were linked through the patient medical record number and the date of surgery. Subjects with no records in the EHR were eliminated. The patient cohort was divided into two datasets: data of patients with surgery from April 2011 to the end of 2012 (model development set) and data of patients with surgery in 2013 (evaluation set). The former dataset was used as the training set for model development. The evaluation dataset was held out fully for the overall evaluation of the developed models. Table 1 describes the detailed demographic information. From April 1, 2011, through December 31, 2013, a total of 6258 procedures with 405 SSIs were collected. The period of April 2011 to the end of 2012 comprised 3996 procedures and 278 SSIs (6.95% rate). About 79% procedures were patients no more than 65 years old, and 21% were patients more than 65 years old. Approximately 83.8% were white, 8.6% were black, and 7.6% were other race/ethnicity and unknown. The year of 2013 comprised 2262 procedures and 127 SSIs (5.6% rate), with similar patient characteristics, as shown in Table 1.
Table 1.
Characteristics of patients and cases of surgical site infection (SSI) from cohorts
| 4/2011–2012 | 2013 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Characteristic | No. of procedure |
No. of SSI |
Superficial SSI |
Deep SSI |
Organ/ Space SSI |
No. of procedure |
No. of SSI |
Superficial SSI |
Deep SSI |
Organ/ Space SSI |
| Total | 3996 | 278 | 140 | 52 | 86 | 2262 | 127 | 47 | 35 | 45 |
| Encounter type | ||||||||||
| Inpatient | 3018 | 265 | 147 | 47 | 83 | 1352 | 108 | 36 | 32 | 40 |
| Outpatient | 978 | 13 | 5 | 5 | 3 | 910 | 19 | 11 | 3 | 5 |
| Age group | ||||||||||
| < 65 | 3160 | 210 | 112 | 41 | 67 | 1569 | 90 | 28 | 28 | 34 |
| ≧65 | 836 | 68 | 38 | 11 | 19 | 693 | 37 | 19 | 7 | 11 |
| Gender | ||||||||||
| Male | 1530 | 122 | 56 | 19 | 47 | 974 | 56 | 21 | 15 | 20 |
| Female | 2466 | 156 | 84 | 33 | 39 | 1288 | 71 | 26 | 20 | 25 |
| Race | ||||||||||
| White | 3366 | 226 | 115 | 45 | 66 | 1881 | 110 | 38 | 31 | 41 |
| Black | 269 | 23 | 10 | 5 | 8 | 142 | 8 | 4 | 1 | 3 |
| Other | 361 | 29 | 15 | 2 | 9 | 239 | 9 | 5 | 3 | 1 |
The clinical data utilized included six data types: demographics, diagnosis codes, orders, lab results, vital signs, and medications. Demographics included each patient’s gender, race, and age at the time of surgery. Diagnosis codes consisted of related ICD-9 codes generated during the encounter and hospital stay at the time of surgery from coding, as well as the diagnoses from the past medical history and problem list. Orders related with SSI diagnosis and treatment were also gathered from the EHR, including imaging orders, infectious disease consultation orders, and procedures with incision and drainage. The most recent lab values and vitals results before surgery and those during the postoperative 30-day window (since surgical adverse events defined as occurring within 30 days after surgery) were collected. Medications utilized for this analysis included antibiotics from the third day after surgery onwards.
Another two important data measures included were American Society of Anesthesiologists (ASA) physical status classification and surgical wound classification. ASA classification from 1 to 6 indicates a patient’s status from normal healthy to declared brain-dead; the surgical wound classification is used for postoperatively grading of the extent of microbial contamination, indicating the chance a patient will develop an infection at the surgical site. We dichotomized the wound classification as the bottom two (no or mild disturbance) versus the remaining levels (significant disturbance).
Data Preprocessing
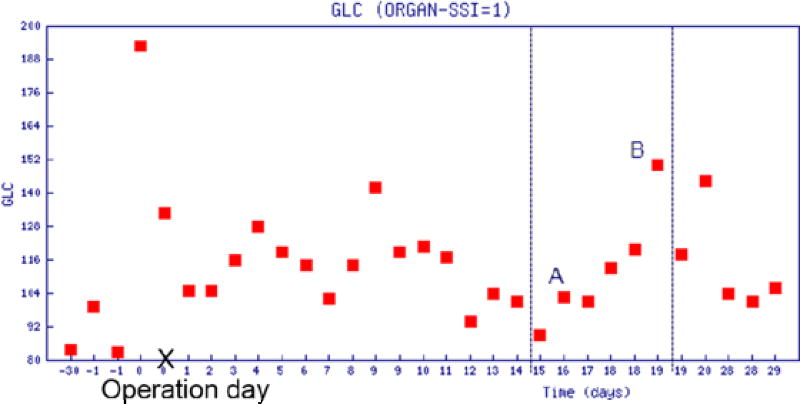
EHR data of interest were collected, cleaned, and analyzed next. Identifying and removing outliers, and correcting inconsistent data were the very first tasks of data preprocessing. How to transform clinical data into meaningful features was our main interest. Most clinical data, such as lab test results and vitals, tended to be longitudinal with repeated measures. Traditional methods to summarize those variables by calculating the moments (mean and standard deviation) or extremes tended not to be sufficient to describe the temporal behavior of such variables. To better summarize individual tests, we explored other features like the change of values during an “elevating period”. An elevating period is a time period during which the measurement in question is near-monotonously increasing from a low level (trough point) to a high level (peak point). For patients with SSI, some lab results, like serum glucose (GLC), platelet count (PLT), and white blood cells (WBC), have significant increases in the measurement from the third day after operation.
As shown in Figure 2, GLC increased in three time periods: (I) day 3~7, GLC increased from 116 to 128; (II) day 7~9, from 104 to 140; and (III) day 15~28, from 87 to 148. Such elevation may indicate the onset of SSI. To capture the elevating period, a feature defined as the postoperative increase from a trough to its nearest peak was included in our tentative model. In the case of multiple elevating periods, the feature was computed by using the period with the highest peak. For measures where low values could indicate SSI, a “descending period” can be defined analogously.
Figure 2.
GLC values within 30 days before and after surgery
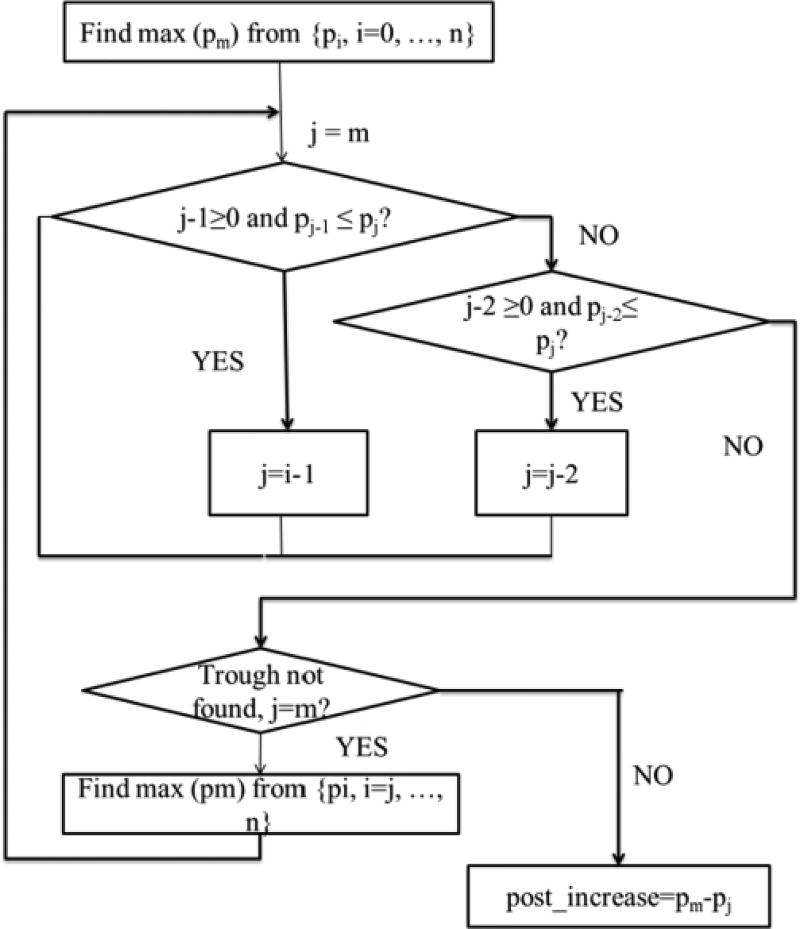
Figure 3 depicts the flowchart of the algorithm to compute this feature. The algorithm first searches for the maximum value (pm) from all results at least two days after the operation ({pi, i=0, …,n}), (e.g., in Figure 2, point B is the maximum GLC value, which was measured nineteen days after the operation). Then the algorithm proceeds by searching for the trough point backward from point B. The algorithm is robust in its filtering of the abnormal point that temporarily breaks the rule of monotone. For example, in Figure 2, the elevating period is from day 15 to 19, however, there is an abnormal point A which breaks the monotone increasing trend between day 15 and 17; to overcome the problem and identify the real trough, the algorithm further compares day 15 and day 17 in order to determine whether the criterion of monotone increasing is satisfied.
Figure 3.
Finding the postoperative increase in GLC
For other data like antibiotic use and specific orders, we created binary variables to indicate whether a relevant element was observed. For example, a value of 1 for Interventional Radiology signifies that an abscess drainage order was placed for a patient; while a value of 0 signifies that no such test was ordered.
Model Development
To build our SSI detection model, we utilized multivariate logistic regression models. We constructed one model for total SSI and one model for each of the three SSI subtypes. Binary variables were entered as dummy indicator variables and continuous variables were entered unmodified. We used stepwise construction to select significant features and Akaike Information Criterion (AIC) for model selection.
Evaluation
In assessing detection of surgical adverse event outcomes like SSI, since these events are relatively rare, overall detection accuracy percentage is not an optimal criterion for evaluating model validity. Instead, we report specificity, as well as the the area under the curve (AUC), in evaluation of our automated detection system. Our aim was to maximize the specificity under the constraint that the negative predictive value remains above 98%. This aim is reflective of our original expectation of actual use of the detection models: to assist a NSQIP chart extractor to eliminate patients who clearly did not suffer the adverse event and then to accelerate the process of data abstraction from clinical charts.
Results
Significant Variables Selected
Tables 2 through 5 show the results for the multivariate detection models for the three kinds of SSI and the total SSI, selected by AIC. The two most common variables included were diagnosis codes (the ICD-9 codes of SSI is 998.xx) and antibiotic use. Superficial SSI occurs just at the skin incision and thus relatively easily diagnosed. Therefore, imaging diagnostic orders tend to be unnecessary. Infection is sometimes diagnosed with microbiology cultures, however, frequently this diagnosis is based on the physical examination only. Actually only cultures ordered or not is a signal of SSI. According to table 3 and table 4, we can find that abscess culture, fluid culture and wound culture are significant factors for detecting deep and organ/space SSI. Since these two kinds of SSIs occur deep within or under the wound, imaging orders for both diagnosis and treatment are frequently required.
Table 2.
Significant indicators for detecting superficial SSI
| Significant variables | Estimate | P-value |
|---|---|---|
| Diagnosis codes | 2.1126 | <0.0001 |
| Wound culture ordered | 2.1941 | <0.0001 |
| Antibiotic use | 1.1321 | <0.0001 |
| Encounter type (inpatient) | 1.6007 | 0.0010 |
| ASA Classification (significant disturbance) | 0.4342 | 0.0058 |
| Abscess culture ordered | 1.5020 | 0.0050 |
| Postoperative increase of GLC | 0.0112 | 0.0687 |
Table 5.
Significant indicators for detecting total SSI
| Significant Variables | Estimate | p-value |
|---|---|---|
| Diagnosis codes | 5.3940 | <0.0001 |
| Antibiotic use | 1.3672 | <0.0001 |
| Abscess culture ordered | 3.2565 | <0.0001 |
| Wound culture ordered | 2.2926 | <0.0001 |
| Imaging diagnosis ordered | 0.8741 | <0.0001 |
| Fluid culture ordered | 1.2909 | <0.0001 |
| Encounter type (inpatient) | 1.0185 | 0.0037 |
| ASA Classification (significant disturbance) | 0.4258 | 0.0031 |
| Preoperative PLT | 0.00214 | 0.0440 |
| Post maximum pain | 0.0775 | 0.0957 |
Table 3.
Significant indicators for detecting deep SSI
| Significant Variables | Estimate | p-value |
|---|---|---|
| Diagnosis codes | 3.1959 | <0.0001 |
| Antibiotic Use | 2.2276 | <0.0001 |
| Abscess culture ordered | 1.2880 | 0.0868 |
| Gram stain ordered | 0.8040 | 0.0427 |
| Imaging treatment ordered | 1.5445 | 0.1107 |
| Imaging diagnosis ordered | 0.6254 | 0.0981 |
| Tissue culture ordered | 1.6516 | 0.1010 |
Table 4.
Significant indicators for detecting organ/space SSI
| Significant Variables | Estimate | p-value |
|---|---|---|
| Imaging treatment | 1.3999 | <0.0001 |
| Imaging diagnosis | 1.2090 | <0.0001 |
| Antibiotic Use | 1.1662 | <0.0001 |
| Abscess culture ordered | 2.3041 | <0.0001 |
| Fluid culture ordered | 1.4204 | 0.0003 |
| Preoperative PLT | 0.00332 | 0.0135 |
| Drainage culture ordered | 1.3760 | 0.0711 |
| Diagnosis code | 0.8259 | 0.0667 |
| Postoperative increase of PLT | 0.0115 | 0.0606 |
We also found the postoperative elevating period of GLC, for superficial and PLT for organ/space, to be indicative of clinical suspicion. Clinically these lab values can be altered in the setting of infection. For a unit increase in postoperative increase of GLC, we expect to see approximately a 0.0112 increase in log-odds of superficial SSI. Similarly, for a unit postoperative increase of PLT, approximately a 0.0115 increase in the log-odds of organ/space SSI is expected.
Model Performance
Four detection models exhibited excellent specificity to eliminate the majority of non-SSI patients, which greatly accelerate the process of extracting postoperative SSI occurrences. Table 6 presents the negative predictive value (NPV) for each of the SSI identification models. The highest specificity 0.988 was for detecting deep SSI at NPV equals to 0.99, and the lowest 0.787 was for detecting total SSI at NPV equals to 0.99. AUC values for the four models were 0.820, 0.898, 0.886 and 0.896, respectively.
Table 6.
Negative predictive value and specificity for four SSI models
| NPV | Specificity | |
|---|---|---|
| Superficial SSI | 0.980 | 1.000 |
| 0.985 | 0.987 | |
| 0.990 | 0.900 | |
|
| ||
| Deep SSI | 0.980 | 1.000 |
| 0.985 | 1.000 | |
| 0.990 | 0.988 | |
|
| ||
| Organ/space SSI | 0.980 | 1.000 |
| 0.985 | 0.999 | |
| 0.990 | 0.974 | |
|
| ||
| Total SSI | 0.980 | 0.935 |
| 0.985 | 0.888 | |
| 0.990 | 0.787 | |
Discussion
The current research is a pilot study to examine the feasibility of automatically detecting postoperative SSI occurrences based on EHR data. The aim of this study is to assist a NSQIP SCR to eliminate patients who clearly did not suffer the adverse event. Therefore, a very high NPV is desired, which could assist in the reliable identification of patients without postoperative SSI. From the modeling results, we can see that all four models perform very well (with specificity ranging from 0.788 to 0.988) in eliminating the majority of patients without SSI based on the NPV equals to 0.99. Considering the nature of NSQIP SCR’s work, SCRs still need to review all clinical charts, even if the positive predictive value for a patient is 0.9 or higher, since they need to extract the clinical characteristics of patients with SSI. Therefore, achieving high NPV, and thus allowing SCRs to eliminate patients, rather than achieving a high positive predictive value, is the main focus of this research.
Among selected potential indicators, a few of them were found to be quite significant with very small p-values. Only the indicators that had p-value less than 0.0001 were employed in the logistic regression modeling, however, this did not improve the detection performance. Other modeling methods, like Random Forest and Support Vector Machine, were employed; however, logistic regression models were found to outperform these methods for detection of all types of postoperative SSI events.
The current study was limited by the fact that it was conducted with only complete cases over three years. This may have limited our ability to fully refine and optimize the automated detection model. In the future, more procedures will be included, and the treatment of missing data will be studied.
Large quantities of meaningful information are stored at the clinical notes, such as imaging reports and culture results, which we did not utilize in this study. For example, a positive abscess culture result could be recorded as “On day 2, isolated in broth only: Bacteroides fragilis group”. However, we merely considered whether the diagnostic and therapeutic imaging orders or cultures were placed, we did not use the actual results. Natural language processing (NLP) has played an important role in detecting adverse events [11–12]. In our future research, we will apply NLP techniques to extract additional important information from clinical notes.
Conclusion
In this study, to accelerate the process of extracting postoperative SSI outcomes from medical charts and reduce the workload of NSIQP SCR, an automated postoperative SSI detection model based on supervised learning was proposed and validated. The models exhibited good performance, they reduced the SCR’s burden by reliably eliminating the vast majority of patients with no SSI. The significant factors of detecting SSI identified by our models are in line with clinical knowledge. In addition, some useful patterns, (e.g. postoperative increase of PLT and GLC), were extracted from the longitudinal lab results.
Acknowledgments
This research was supported by the University of Minnesota Academic Health Center Faculty Development Award (GS, GM) and the American Surgical Association Foundation (GM). The authors thank Fairview Health Services for support of this research.
References
- 1.Fetter RB, Shin Y, Freeman JL, Averill RF, Thompson JD. Casemix definition by diagnosis-related groups. Med Care. 1980;18(2 Suppl):1–53. [PubMed] [Google Scholar]
- 2.Dimick JB, Osborne NH, Hall BL, Ko CY, Birkmeyer JD. Risk-adjustment for comparing hospital quality with surgery: How many variables are needed? J Am Coll Surg. 2010 Apr;210(4):503–508. doi: 10.1016/j.jamcollsurg.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guillamondegui OD, Gunter OL, Hines L, Martin BJ, Gibson W, Chris Clarke P, Cecil WT, Cofer JB. Using the National Surgical Quality Improvement Program and the Tennessee Surgical Quality Collaborative to Improve Surgical Outcomes. J Am Coll Srug. 2012 Apr;2014(4):709–14. doi: 10.1016/j.jamcollsurg.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Rutala WA, Weber DJ. Cleaning, disinfection, and sterilization in healthcare facilities: What clinics need to know. Clin Infect Dis. 2004 Sep 1;39(5):702–9. doi: 10.1086/423182. [DOI] [PubMed] [Google Scholar]
- 5.http://www.cdc.gov/nhsn/acute-care-hospital/ssi/
- 6.http://site.acsnsqip.org/wp-content/uploads/2012/03/ACSNSQIP-Participant-User-Data-File-UserGuide_06.pdf.
- 7.Mu Y, Edwards JR, Horan TC, Berrios-Torres SI, Fridkin SK. Improving risk-adjusted measures of surgical site infection for the national healthcare safety network. Infect Control Hosp Epidemiol. 2011 Oct;32(10):970–86. doi: 10.1086/662016. [DOI] [PubMed] [Google Scholar]
- 8.Levine PJ, Elman MR, Kullar R, Townes JM, Bearden DT, Vilches-Tran R, McClellan I, McGregor JC. Use of electronic health record data to identify skin and soft tissue infections in primary care settings: a validation study. BMC Infectious Disease. 2013 Apr 10;13:171. doi: 10.1186/1471-2334-13-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warren DK, Nickel KB, Wallace AE, Mines D, Frasesr VJ, Olsen MA. Can additional information be obtained from claims data to support surgical site infection diagnosis codes? Infect Control Hosp Epidemiol. 2014 Oct;35(Suppl 3):S124–32. doi: 10.1086/677830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price CS, Satitz LA. Final report. Rockville, MD: Agency for Healthcare Research and Quality; Mar, 2012. Improving the measurement of surgical site infection risk stratification/outcome detection. (prepared by Denve health and its partners under contract No. 290-2006-00-20). AHRQ publication No. 12-0046-EF. [Google Scholar]
- 11.Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc. 2005 Jul-Aug;12(4):448–57. doi: 10.1197/jamia.M1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bates DW, Evans RS, Murff H, Stetson PD, Pizziferri L, Hripcsak G. Detecting adverse events using information technology. J Am med Inform Assoc. 2003 Mar-Apr;10(2):115–28. doi: 10.1197/jamia.M1074. [DOI] [PMC free article] [PubMed] [Google Scholar]