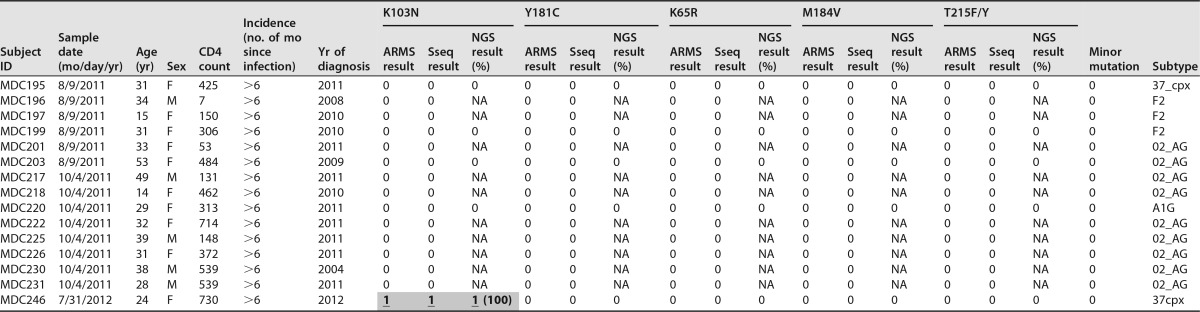
TABLE 3.
Characteristics of study subjects and comparative HIVDR mutation profiles for 5 mutations using 3 testing methodsa
Detected drug resistance mutations are shaded in gray. “0” indicates the absence of the respective drug resistance mutation (<5% of quasispecies; not considered to be significant), and “1” indicates the presence of the respective drug resistance mutation (≥5% of quasispecies). ARMS, ARMS-PCR; Sseq, Sanger sequencing; NGS, next-generation sequencing; F, female; M, male; NA, not applicable. Shaded underlined boldface numbers indicate calls positive by all 3 testing methods, and shaded italic numbers indicate false-positive calls by ARMS-PCR and NGS for minorities. Minor HIVDR mutations are indicated if present. The HIV subtypes were determined with HIV BLAST (LANL database) and phylogenetic analysis of the reverse transcriptase sequences generated by standard sequencing and NGS (Fig. 1).
b MDC192 is either subtype CRF02_AG or subtype CRF36_cpx.