ABSTRACT
Western blotting (WB) for human T cell leukemia virus type 1 (HTLV-1) is performed to confirm anti-HTLV-1 antibodies detected at the initial screening of blood donors and in pregnant women. However, the frequent occurrence of indeterminate results is a problem with this test. We therefore assessed the cause of indeterminate WB results by analyzing HTLV-1 provirus genomic sequences. A quantitative PCR assay measuring HTLV-1 provirus in WB-indeterminate samples revealed that the median proviral load was approximately 100-fold lower than that of WB-positive samples (0.01 versus 0.71 copy/100 cells). Phylogenic analysis of the complete HTLV-1 genomes of WB-indeterminate samples did not identify any specific phylogenetic groups. When we analyzed the nucleotide changes in 19 HTLV-1 isolates from WB-indeterminate samples, we identified 135 single nucleotide substitutions, composed of four types, G to A (29%), C to T (19%), T to C (19%), and A to G (16%). In the most frequent G-to-A substitution, 64% occurred at GG dinucleotides, indicating that APOBEC3G is responsible for mutagenesis in WB-indeterminate samples. Moreover, interestingly, five WB-indeterminate isolates had nonsense mutations in Pol and/or Tax, Env, p12, and p30. These findings suggest that WB-indeterminate carriers have low production of viral antigens because of a combination of a low proviral load and mutations in the provirus, which may interfere with host recognition of HTLV-1 antigens.
KEYWORDS: nonsense mutation, nucleotide substitution, proviral load, provirus, Western blot indeterminate, human T cell leukemia virus, nucleic acid technology
INTRODUCTION
Human T cell leukemia virus type 1 (HTLV-1) infection is endemic in various regions, including sub-Saharan Africa, the Caribbean, parts of South America, the Middle East, Australo-Melanesia, and the southwestern area of Japan (1, 2). HTLV-1 can be transmitted through prolonged breast feeding, sexual intercourse, and transfusion of contaminated blood. In some African countries, zoonotic transmission to humans by severe bites from simian T cell leukemia virus type 1-infected monkeys has been observed (3). The majority of infected people live without any symptoms; however, in a portion of carriers, HTLV-1 causes adult T cell leukemia (ATL), HTLV-1-associated myelopathy/tropical spastic paraparesis, and HTLV-1 uveitis/HTLV-1-associated uveitis after a long period of latency (4).
Diagnosis of HTLV-1 infection is usually made by serological testing at the initial screening of blood donors, in pregnant women, and in suspected HTLV-1-related diseases. A variety of serological screening kits are available, including chemiluminescent enzyme immunoassays (CLEIAs), chemiluminescent immunoassays, and particle agglutination assays, followed by confirmation by Western blotting (WB) (5–9). WB measures the serological reaction to both Gag core proteins (p19, p24, and p53) and the Env protein gp46 (10). In the ProBlot HTLV-1 WB test kit, at least one Gag band and an Env gp46 band should be detected for an HTLV-1-positive result. However, incomplete antibody binding to HTVL-1 Gag or Env is often observed and therefore the result is classified as indeterminate (see Table S1 in the supplemental material).
The proportion of indeterminate WB results is reported to be high in areas such as Zaire (68%) and Central Africa (65.65%) (11, 12). The frequent occurrence of the indeterminate pattern in WB makes it difficult to diagnose the infection correctly. Causes of these indeterminate results have been reported to be cross-reactivity with Plasmodium falciparum infection (13–15), infection with HTLV-3 and HTLV-4 (16, 17), and delayed seroconversion with low antibody titers (18–22). In those with WB-indeterminate samples, the indeterminate result is sometimes sustained for a long time (18, 19, 23). Nevertheless, it has been reported that a significant portion of HTLV-1 WB-indeterminate samples are positive for provirus by DNA testing, i.e., 12.5% of WB-indeterminate blood donors in Iran, 9.2% in Brazil, 14.7% (5 of 34) in Argentina, and 42% of patients with neurologic symptoms and 44% of blood donors in the United States (19, 24–27). Thus, in addition to serological testing, proviral DNA detection by quantitative PCR (qPCR) and/or qualitative PCR with HTLV-1-specific primers and a probe against genomic DNA is considered one of the best methods to resolve issues in diagnosis. However, the reason why HTLV-1 provirus-positive blood initially returns an indeterminate result by WB is unclear, especially as it is unlikely that all provirus-positive samples were in the window period of the viral infection prior to antibody formation.
In this study, we assessed the mechanism causing indeterminate WB results by analyzing the complete HTLV-1 genome in WB-indeterminate samples. Furthermore, we evaluated the advantages of HTLV-1 qPCR for cases that were not clearly diagnosed by serological testing.
RESULTS
Provirus detection in WB-indeterminate samples.
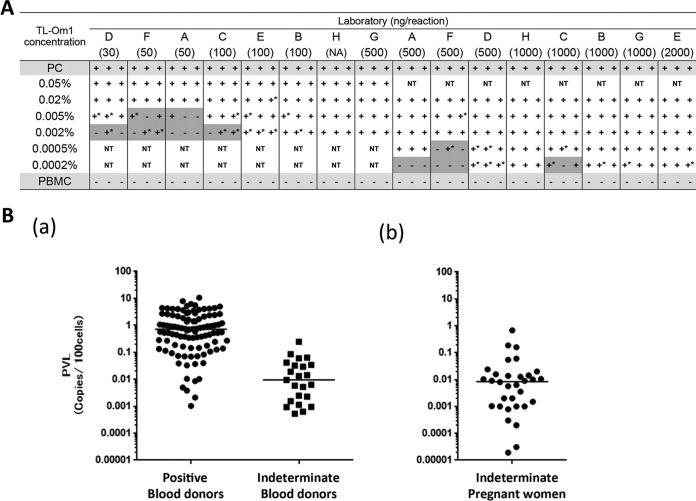
To detect the HTLV-1 provirus with high sensitivity, we first evaluated the suitable amount of genomic DNA used in HTLV-1 qPCR in eight Japanese laboratories. To assess the detection limit of the qPCR assay in these laboratories, peripheral blood mononuclear cells (PBMCs) that were spiked with low concentrations of HTLV-1-infected cells were analyzed. All laboratories could detect the provirus at concentrations of approximately 4 copies/105 cells when laboratories used >500 ng of genomic DNA in the PCR (Fig. 1A).
FIG 1.
PVLs of WB-indeterminate clinical samples. (A) qPCR was performed three times on different days independently with PBMCs spiked with TL-Om1 cells at concentrations of 0.0002 to 0.05%. Laboratories used different amounts of genomic DNA and their in-house qPCR methods. A number in parentheses under a letter corresponding to a laboratory indicates the amount (in nanograms) of DNA used in the reaction mixture. The PC (positive-control) sample consisted of genomic DNA from 0.8% TL-Om1/PBMC. Tests were performed with duplicate or triplicate wells. A plus sign indicates that all of the wells were positive, a minus sign indicates that all of the wells were negative, and a plus sign with an asterisk indicates that there was at least one negative well in the results. NT, not tested. Gray shading indicates that there was at least one negative result in the test. (B, part a) HTLV-1 PVLs (number of copies per 100 cells) of WB-positive (n = 100; left) and WB-indeterminate (n = 23; right) blood donors. (B, part b) PVLs of WB-indeterminate pregnant women (n = 32). Bars indicate median PVLs.
We collected genomic DNA from the peripheral blood of pregnant women with indeterminate WB results (n = 196) from all over Japan and of blood donors from two geographic areas, one where HTLV-1 infection is endemic (n = 39) and the other where it is not (n = 61). The frequency of HTLV-1 provirus and the HTLV-1 proviral loads (PVLs) in these WB-indeterminate samples were then measured by the optimized qPCR method (by using 1 μg of genomic DNA). The percentage of provirus-positive blood donors differed according to where the blood was collected (46.2% where HTLV-1 infection is endemic and 8.2% where it is not) (Table 1). Similarly, provirus was detected in 16.5% of WB-indeterminate pregnant women. Among the provirus-positive samples, the median PVL (number of copies per 100 cells) was 0.011 in blood donors (n = 23) and 0.008 in pregnant women (n = 32) (Fig. 1 and Table 1). Meanwhile, the median PVL of WB-positive blood donors (n = 100, a mixture of the two sample areas) was 0.71 copy/100 cells. From these results, the PVL of WB-indeterminate samples was approximately 100-fold lower than that of WB-positive donors (Fig. 1). The antibody titers of provirus-positive WB-indeterminate samples were higher than those of provirus-negative WB-indeterminate samples (P < 0.0001; Fig. S2). A significant correlation between PVLs and antibody titers in the initial screening test of blood donors was not observed (data not shown).
TABLE 1.
Provirus detection by qPCR and PVLs of WB-indeterminate samples
| Sample source and regiona | Total no. analyzed | No. (%) provirus positive | Median PVLb (95% CIc) |
|---|---|---|---|
| Blood donors | |||
| Endemic | 39 | 18 (46.2) | 0.011 (0.002–0.029) |
| Nonendemic | 61 | 5 (8.2) | |
| Pregnant women nationwide | 194 | 32 (16.5) | 0.008 (0.002–0.014) |
Where HTLV-I infection is endemic or nonendemic.
HTLV-1 PVL (number of copies/100 cells).
CI, confidence interval.
Phylogenetic features of the provirus were not associated with the indeterminate result.
In the HTLV-1 screening, we occasionally had samples that the PCR results indicated were positive for HTLV-1; however, these infections could not be confirmed by WB. For example, Matsumoto et al. recently reported that 33 of 600 CLEIA-positive blood donor samples were provirus positive but WB indeterminate and 2 of 600 CLEIA-positive samples were provirus positive but WB negative (28). We hypothesized that genomic features of HTLV-1 may be associated with the indeterminate results of the antibody test.
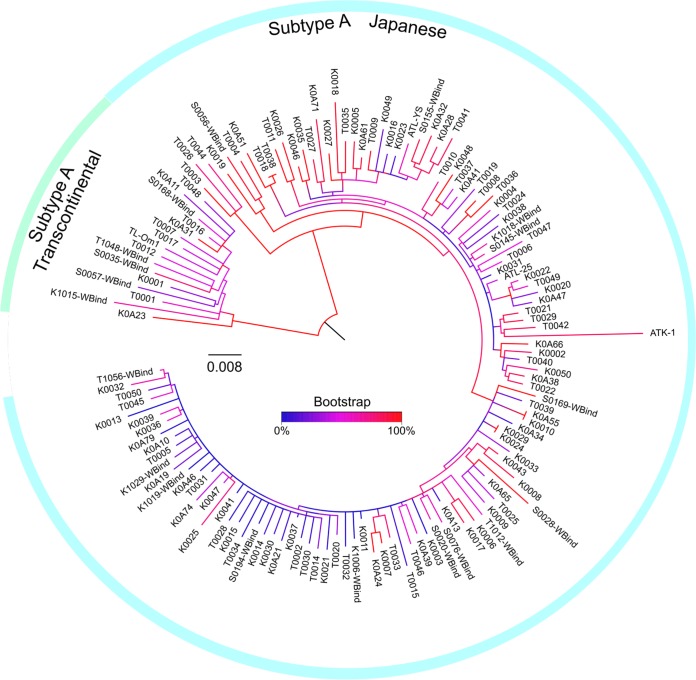
To investigate the causative phylogenetic feature of HTLV-1 in WB-indeterminate blood donor samples, the full genomic sequences of 114 HTLV-1 WB-positive and 19 WB-indeterminate samples were determined by direct sequencing. A total of 1,085 single nucleotide variants (SNVs) were found in these 133 isolates and four HTLV-1 genomes that were registered as from Japan (ATK-1, ATL-YS, ATL-25, and TL-Om1). A phylogenetic tree was drawn (Fig. 2) with RAxML, which utilizes a maximum-likelihood method. The majority of isolates belonged to the subtype A Japanese (JP) subgroup, while a small portion of isolates belonged to the subtype A transcontinental (TC) subgroup. WB-indeterminate isolates were dispersed throughout both the JP and TC branches (Fig. 2 and Table 2). The frequency of TC-type WB-indeterminate samples in the tree appears relatively high compared with that of the JP type (5 of 10 and 14 of 118, respectively); however, it is difficult to compare the frequencies of those two groups statistically because the geographic background of these pregnant women is unknown. Importantly, distinct WB-indeterminate strains that clustered in specific regions of the phylogenetic tree were not found.
FIG 2.
Phylogenetic analysis of HTLV-1 isolates from WB-indeterminate samples. SNVs of 1,085 nucleotide positions in the full HTLV-1 genomes of WB-positive (n = 114) and -indeterminate (n = 19) samples were analyzed by RAxML, an algorithm that uses the maximum-likelihood method. The tree was drawn by using the GTRGAMMA model. WBind indicates an HTLV-1 sequence from a WB-indeterminate sample. ATK-1 (J02029), ATL-YS (HTU19949), ATL-25 (AB513134), and TL-Om1 (AB979451) are complete HTLV-1 sequences from clinical samples or a cell line derived from Japanese donors.
TABLE 2.
Phylogenetic types of HTLV-1 in WB-positive and -indeterminate samples
| Sample source and WB result | Total no. analyzed | No. of HTLV-1 subtype A subgroup: |
|
|---|---|---|---|
| JP | TC | ||
| Blood donors | |||
| Positive | 114 | 104 | 10 |
| Indeterminate | 8 | 6 | 2 |
| Pregnant women, indeterminate | 11 | 8 | 3 |
Characteristics of nucleotide substitutions in the HTLV-1 genomes of WB-indeterminate samples.
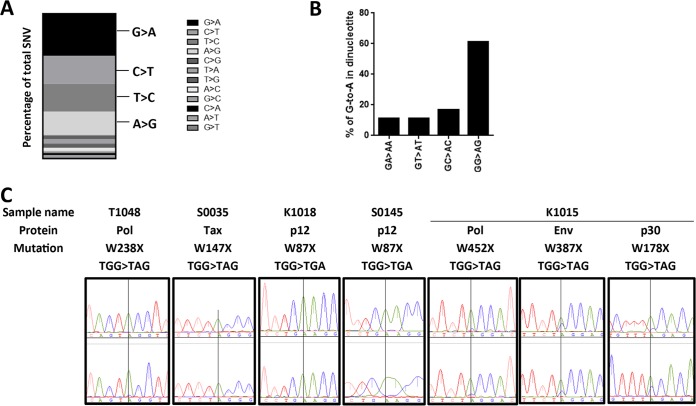
To determine the host enzymes responsible for the mutagenesis of the HTLV-1 genomes in WB-indeterminate samples, such as the APOBEC family, we focused on the nucleotide substitutions in the HTLV-1 genomes. Among the total of 1,085 SNVs found in the HTLV-1 genomes from 114 HTLV-1 WB-positive and 19 WB-indeterminate samples, there were 135 indeterminate WB result-specific single nucleotide substitutions. The most frequent type of substitution was G to A (28.9%), followed by C to T (19.3%), T to C (19.3%), and A to G (16.3%) (Fig. 3A). These four types of substitutions were responsible for 83.8% of the indeterminate WB result-specific substitutions. Moreover, the majority of G-to-A substitutions occurred at GG dinucleotides (64.1%), suggesting that a large portion of these G-to-A substitutions were mediated by APOBEC3G (29) (Fig. 3B).
FIG 3.
Characteristics of nucleotide substitutions in WB-indeterminate samples. (A) The occurrence of types of nucleotide substitutions is represented as a percentage of the total number of mutations. The four major substitutions, G-to-A, C-to-T, T-to-C, and A-to-G mutations, are indicated. (B) Percentages of second-nucleotide use at G-to-A mutation sites. (C) Electropherogram of stop codon substitutions. Two representative electropherograms per sample are shown. The nucleotide with a vertical line through it is the position of the G-to-A substitution.
Characteristics of HTLV-1 genomic sequences in WB-indeterminate samples.
We then focused on the mutations associated with viral replication in HTLV-1 from WB-indeterminate samples. To our surprise, among the 19 full HTLV-1 genomic sequences from WB-indeterminate samples, five isolates had nonsense mutations in the coding region of viral proteins, namely, two in Pol, one in Env, one in Tax, two in p12, and one in the p30 sequence (Table 3). Because p30 and p13 use the same coding frame, a p30 W178X mutation (sample K1015) also leads to a p13 W24X mutation. Thus, these HTLV-1 isolates apparently have a fatal replication defect. As shown in Fig. 3C, almost all of the peaks of the electropherograms of nucleotide sequencing of these stop codon mutations were a single peak, suggesting that the HTLV-1 clones with abortive replication constitute the major clone or clones in the carrier. Interestingly, the nonsense mutations in K1015 were mixed with wild-type sequences. In contrast, among the HTLV-1 genomes of 114 WB-positive blood donors, there was only one premature termination in the C-terminal region of Tax (Q334X, accession no. LC209961) and one premature termination in the C-terminal region of p12 (W82X, accession no. LC210033). There were no other premature terminations in the WB-positive samples. This indicated that these genomic mutations with premature termination are possibly a specific feature of WB-indeterminate samples. In addition, all WB-indeterminate samples had many unique mutations in a variety of proteins (Table 4). Including the nonsense mutations in Table 3, on average, there were 4.7 amino acid changes in the provirus of WB-indeterminate samples. Among the unique mutations, there were mutations that cause amino acid charge changes, including Asp, Glu, Arg, and Lys mutations (36%, 30 of 83), and cause structure changes such as a Pro mutation (14%, 12 of 83). These accumulated mutations suggest that there are dramatic changes in the function of viral proteins that may lead to decreased HTLV-1 replication efficiency.
TABLE 3.
Abortive genetic changes in HTLV-1 genes in WB-indeterminate samples
| Sample source and name | PVLa | Amino acid mutation(s) in viral protein: |
Phenotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gag | Pro | Pol | Env | Rex | Tax | p12 | p30 (p13) | HBZ | |||
| Blood donors | |||||||||||
| T1048 | 0.013 | W238X | Premature termination | ||||||||
| K1015 | 0.244 | W452X | W387X | W178X (W24X) | Premature termination | ||||||
| K1018 | 0.060 | W87X | Premature termination | ||||||||
| Pregnant women | |||||||||||
| S0035 | 0.006 | W147X | Premature termination | ||||||||
| S0145 | 0.677 | W87X | Premature termination | ||||||||
HTLV-I PVL (number of copies per 100 cells).
TABLE 4.
Genetic changes with unknown significance in HTLV-1 genes in 19 WB-indeterminate samples
| Sample source and name | PVLb | Amino acid mutation(s) in viral protein: |
Nucleotide mutation(s) in LTR | Phenotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gag | Pro | Pol | Env | Rex | Tax | p12 | p30 | HBZ | ||||
| Blood donors | ||||||||||||
| T1012 | 0.019 | E169D | K93R, T557A | G460R | S134F | Unknown | ||||||
| T1048a | 0.013 | P100S | F54L | 490T>G, 559C>T | ||||||||
| T1056 | 0.032 | M701V | H347TfsX18 | 53_54insA, 116_117insC | Tax frameshift | |||||||
| K1006 | 0.034 | T345N | V16A | G52E, T62M | G191R, R201C | Unknown | ||||||
| K1015a | 0.244 | A9T | V293I, S355C, G482R | V247A, G446R | V29A | G21R, G137R, G259R | S168P | 544C>T | ||||
| K1018a | 0.060 | Q46E | H503R | T267del | G90E | P63S | ||||||
| K1019 | 0.084 | G850S | A264V | R7K, S70N | A209T | Unknown | ||||||
| K1029 | 0.009 | A156V, V161I | 479delT, 289C>T | Unknown | ||||||||
| Pregnant women | ||||||||||||
| S0020 | 0.160 | Q206R | L64P | F67L | K87R | 239T>C | Unknown | |||||
| S0028 | 0.006 | R166M | I204V | G96D, E179D | G65S, N185Y | R222Q | 355G>A | Unknown | ||||
| S0035a | 0.006 | S113P, P409S | C183Y, G188R | R12Q, G183R | P8S, G166R | 319G>A | ||||||
| S0056 | 0.014 | L267R | T4A | F62L | 73A>T, 146C>T | Unknown | ||||||
| S0057 | 0.016 | R259K, I433M | G144D, C162Y | P23S | L26F | R127K | 618A>G | Unknown | ||||
| S0076 | 0.008 | P547S | H347TfsX18 | F118S | K35E | 53_54insA | Tax frameshift | |||||
| S0145a | 0.677 | E173K | G90E | P63S | 487T>C | |||||||
| S0155 | 0.024 | S162F | Y678C | T151A | 353G>A | Unknown | ||||||
| S0168 | 0.055 | Q812R | G29S | 585T>A, 744A>G | Unknown | |||||||
| S0169 | 0.014 | L17F | K855R | L164R | Unknown | |||||||
| S0194 | 0.059 | P103_S104insPP | S81P | 587T>C | Unknown | |||||||
Sample also has abortive mutations.
HTLV-1 PVL (number of copies/100 cells).
DISCUSSION
We successfully established the HTLV-1 qPCR assay for analysis of the PVLs of WB-indeterminate samples after initially estimating the detection limit of the HTLV-1 qPCR assay. An important feature of HTLV-1 discovered in WB-indeterminate samples was that the PVL of WB-indeterminate samples was generally extremely low. The PVL was approximately 100-fold lower than that of carriers with WB-positive results. Interestingly, there was a geographic difference between the provirus DNA positivity rates of WB-indeterminate blood donors in areas where HTLV-1 infection is not endemic and areas where it is, 8.2 and 46.2%, respectively (Table 1). These rates were as high as those previously reported in other areas of the world, including Iran and the United States. The geographic changes may be produced by the balance of the population of indeterminate samples from the true negative (HTLV-1-uninfected) group, which were originally identified as background in the WB test, and indeterminate samples from the true positive (HTLV-1-infected) group, which were not able to be identified as positive by the WB test. Therefore, the PCR positivity rate will rise in areas where HTLV-1 infection is endemic because the higher number of indeterminate WB results from true positive is increased.
Using more than 100 complete HTLV-1 genome sequences from areas where HTLV-1 infection is endemic and where it is not, we produced an overview of the phylogenic tree of Japanese HTLV-1. Importantly, by adding the HTLV-1 sequences of WB-indeterminate samples to the tree, we revealed that there were no specific subgroups of strains that frequently generate indeterminate WB results in Japan. In other words, one of the causes of indeterminate WB results may be associated with individual HTLV-1 nucleotide mutations rather than the strain of HTLV-1. Although our results are applicable to WB-indeterminate samples from Japanese carriers, the cause of indeterminate WB results in other HTLV-1 strains around the world will be elucidated by precise genomic analysis in further studies. These results may also be useful for the improvement of HTLV-1 diagnostic kits.
It has been reported that HTLV-related viruses or malaria infection cause indeterminate WB results; however, these causes are applicable only in limited areas of the world and Japan is not an area where these pathogens are endemic (13, 15–17). We assessed the cause of indeterminate HTLV-1 WB results by analysis of the entire genomic sequence of HTLV-1 and found that a portion of HTLV-1 strains with indeterminate results have a premature termination codon in viral proteins. These mutations apparently decreased the virus's replication efficiency because the viral proteins could not function like those of the wild-type virus, which possibly led to decreased antigen expression in the long term. We think the mechanism of emergence of these mutated proviruses is that in WB-indeterminate carriers, wild-type virus-infected cells have been eliminated by the host immune system and eventually only mutated viruses with low antigen production remain. This hypothesis is supported by our finding that there remained a faint wild-type sequence in some electropherograms with nonsense mutations (Fig. 3C). Abortive HIV-1 infection was reported previously in samples with indeterminate WB results (30). In addition, a report showed Tax point mutations in HTLV-1 WB-indeterminate samples (31). In our study, premature stop codons were observed not only in Tax but also in various HTLV-1 coding regions, such as Pol, Env, p12, and p30, in WB-indeterminate samples, indicating an association of abortive HTLV-1 strains with indeterminate WB results. In addition, unique mutations were observed not only in the target proteins of WB tests such as Gag and Env but also other HTLV-1 proteins. Interestingly, the Tax G137R mutation was observed in K1015. This amino acid is critical for the Tax function of NF-κB signaling (32). Furthermore, Rex T4A (S0056) and P8S (S0035) mutations are located in the RxRE association domain and the Rex T62M (K1006) mutation is located in the Rex multimerization domain (33). This leads us to hypothesize that the cause of indeterminate results is not only the inadequate sensitivity of the diagnostic kit for Env and Gag antibody detection but also the nature of HTLV-1 in WB-indeterminate samples. Mutated HIV-1 can revert back to the wild type after transmission (34, 35). However, we believe that this is not the case in HTLV-1 of WB-indeterminate samples. Generally, after settlement of HTLV-1 infection, HTLV-1 prefers to disseminate through mitotic division of infected cells with cellular DNA polymerase (36). Moreover, PVLs are low in WB-indeterminate samples and replication-incompetent mutations are dominant in a portion of HTLV-1 of WB-indeterminate samples. Thus, HTLV-1 in WB-indeterminate samples almost lost the opportunity to introduce mutations back into the wild type by HTLV-1 reverse transcriptase at transmission.
To summarize our results, WB-indeterminate samples could be divided into four groups on the basis of their PVLs and genomic features (Table 5). The first group is negative for HTLV-1 provirus. This includes true-negative samples and those undetectable by qPCR. In the second group, HTLV-1 provirus is detected despite the extremely low PVL. This possibly includes wild-type HTLV-1 provirus. In the third group, the abortive HTLV-1 strain is dominant. In the fourth group, unique amino acid or nucleotide mutations are present in the provirus. A common property of the provirus of the latter three groups of provirus-positive samples would be an extremely low level of HTLV-1 antigen production. Thus, we could conclude that WB-indeterminate samples tend to have an extremely low level of HTLV-1 antigen expression because of specific features of the HTLV-1 genome. This low antigen level leads to an insufficient antibody titer for the determination of infection by WB. Sustained indeterminate WB results over a prolonged period could be partially explained by this hypothesis (18–20, 23).
TABLE 5.
Summary of proviral features of WB-indeterminate samples in this study
| No. of samplesa analyzed | qPCR result | HTLV-1 genome | Type of mutation |
|---|---|---|---|
| 239 | Provirus negative | ||
| 36 | Provirus positive | Not determined | |
| 14 | Determined | Unique | |
| 5 | Nonsense |
Total of 294.
Interestingly, APOBEC3G, a host mediator of GG-to-AG substitutions, facilitates the abortion of viral, including HIV-1, replication (37). However, in our study, its function possibly facilitates the survival of HTLV-1 provirus through the decreased production of viral antigens, leading to escape from the host immune system. We think this fits well with the explanation for the reports of the frequent PCR positivity of WB-indeterminate samples (19, 24, 26). Fan et al. analyzed mutations in ATL and reported that among the mutations in ATL, G-to-A is the most frequent and a GG-to-AG substitution was also prominent in all G-to-A mutations (29). They additionally showed the frequent occurrence of stop codon substitutions in the HTLV-1 genome in ATL. Our findings on the mutation status of WB-indeterminate samples are thus in accordance with those reported for ATL. The reason why the same phenomena were observed in the HTLV-1 genome in both ATL and WB-indeterminate samples is unknown. It will be further elucidated through the precise analysis of strategies used by HTLV-1 to continue to reside in carriers.
Finally, our finding that the provirus exists with reduced replication activity in a portion of WB-indeterminate carriers through genetic mutation in the provirus strongly emphasizes the importance of nucleotide amplification testing, such as qPCR, for the diagnosis of HTLV-1 infection.
MATERIALS AND METHODS
Cells and clinical samples.
TL-Om1 cells were a kind gift from Kazuo Sugamura (Miyagi Cancer Center Research Institute) (38). TL-Om1 cells were maintained in RPMI 1640 (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin-streptomycin (Invitrogen, Carlsbad, CA, USA), 2 mmol/liter l-glutamine, and 10 ng/ml interleukin-2 (PeproTech, London, United Kingdom). PBMCs were purchased from AllCells Inc. (Alameda, CA, USA). Cryopreserved PBMCs were resuspended in RPMI 1640 supplemented with 10% FBS at 37°C in accordance with the protocol provided by AllCells Inc.
PBMCs from HTLV-1 WB-indeterminate pregnant women were obtained with informed consent. Blood clots from HTLV-1 WB-indeterminate blood donors were obtained in two different areas of HTLV-1 epidemiology, the Kanto Block Blood Center, in an area where HTLV-1 infection is not endemic, including the prefectures of Tokyo and Chiba, and the Kyushu Block Blood Center, in an area where HTLV-1 infection is not endemic, including the prefectures of Kyushu Island. The kit used for initial blood donor screening was Lumipulse Presto HTLV-I (Fujirebio, Tokyo, Japan), one of the CLEIAs. The WB kit used for confirmation of the first screening results was ProBlot HTLV-I (Fujirebio, Tokyo, Japan) (7). Briefly, in ProBlot HTLV-I, bands of p19, p24, and p53 for Gag and gp46 for Env are used for interpretation of the result. The bands were defined by three grades, namely, −, ±, and +. If all bands are −, the result is judged as negative. When Env gp46 and Gag p19, p24, or p53 are +, the result is judged as positive. Band patterns that are neither negative nor positive are judged as indeterminate (Table S1). The antibody titers and profiles of WB-indeterminate patterns of blood donors are listed in Table S2. Information about the kinds of kits used for initial screening of pregnant women was unavailable. In addition, the antibody titers and WB band patterns of pregnant women were unavailable. This study was approved by the ethical review boards of the National Institute of Infectious Diseases (Institutional Review Board approval no. 392).
Eight Japanese laboratories (one national institute [the National Institute of Infectious Diseases], five universities [The University of Tokyo, the St. Marianna University School of Medicine, Nagasaki University, the University of Miyazaki, and Kagoshima University], one Japanese Red Cross laboratory [the Central Blood Institute], and one diagnostic test company [SRL Inc.]) participated in this study.
Preparation of HTLV-1 cell dilutions.
Previously, we found that the HTLV-1 copy number of the TL-Om1 genome was 1.8/cell by fluorescence in situ hybridization analysis (39). The method used to stain TL-Om1 cells with carboxyfluorescein diacetate succinimidyl ester (CFSE) was previously described (40). CFSE-stained TL-Om1 cells that were resuspended in Cellbanker (TaKaRa Bio, Osaka, Japan) were serially diluted with PBMCs at the concentrations described in Fig. 1 and frozen at −80°C. The concentrations of TL-Om1 cells were analyzed by flow cytometry with a JSAN flow cytometer (Bay Bioscience, Kobe, Japan) (Tables S3 and S4). A series of the same frozen samples packed in dry ice were then provided to the participating laboratories by the National Institute of Infectious Diseases.
Estimation of detection limit of HTLV-1 qPCR.
The DNA extraction methods of the laboratories have been described previously (40). The protocols for HTLV-1 qPCR performed in the eight laboratories have also been reported previously (41–47) (Table S5).
HTLV-1 qPCR was performed with purified DNA in laboratories independently three times on different days. To evaluate all of the preparation steps, each measurement began with the extraction of genomic DNA from aliquots of frozen cell samples provided to each laboratory and testing was performed once with the extracted DNA.
Analysis of HTLV-1 genomic sequences.
The full-length genomic sequence of HTLV-1 was amplified from four regions by long PCR with the KOD-FX neo polymerase kit (Toyobo, Tokyo, Japan) in accordance with the manufacturer's protocol. PCR products were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany). The sequencing PCR was performed with the BigDye ver 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) with sequencing primers in accordance with the manufacturer's protocol. All of the primer sequences used in the long PCR and the sequencing PCR are described in Fig. S1 and Tables S6 and S7. The sequences of PCR products were read from both strands. For sequencing of the GC-rich region of the HTLV-1 genome, equivalent to nucleotides 2099 to 2124 of the ATK-1 (accession no. J02029) reference strain, the dGTP BigDye Terminator v3.0 kit (Applied Biosystems) was used with primers 34F (GGAGATATGTTGCGGGCTTGT) and 41R (GGGAGGTGAGCTTAAAGTGATCTT), respectively, in accordance with the kit protocol. The sequence was determined with an Applied Biosystems 3730 DNA analyzer. Contigs were composed by the sequence assembling software ATGC (GENETYX, Tokyo, Japan). Complete long terminal repeat (LTR) sequences were determined by combining consensus regions of 5′ and 3′ LTR reads as described previously (48). HTLV-1 sequences of T0018 and T0038 were obtained from the same donor on different donation dates.
Phylogenetic analysis.
In addition to blood donor samples in which the PVLs were quantitated, a further 23 genomic sequences of HTLV-1 from WB-positive blood donors from the Kyushu Block Blood Center were added to the phylogenetic analysis. For phylogenetic analysis, SNVs were extracted and analyzed with RAxML by the maximum-likelihood method with 1,000 bootstrap samples. The phylogenetic tree was inferred by using the GTRGAMMA model.
Accession number(s).
The HTLV-1 nucleotide sequences of WB-indeterminate samples have been submitted to the DNA Data Bank of Japan (DDBJ) and assigned NCBI accession numbers LC185235 to LC185242 and LC192254 to LC192264. The HTLV-1 sequences of WB-positive samples have also been submitted to the DDBJ and assigned NCBI accession numbers LC209958 to LC210071.
Supplementary Material
ACKNOWLEDGMENTS
We thank Isao Naruse, Setsuko Sato, Takuo Mizukami, Shuji Izumo, and Shimeru Kamihira for their expert assistance.
This work was supported by the Ministry of Health, Labour and Welfare (MHLW) (H23-sinkou-ippan-016) and the Research Program on Emerging and Re-emerging Infectious Diseases of the Japan Agency for Medical Research and Development, AMED (H26-sinkoujitsuyouka-ippan-013).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00659-17.
REFERENCES
- 1.Satake M, Yamaguchi K, Tadokoro K. 2012. Current prevalence of HTLV-1 in Japan as determined by screening of blood donors. J Med Virol 84:327–335. doi: 10.1002/jmv.23181. [DOI] [PubMed] [Google Scholar]
- 2.Gessain A, Cassar O. 2012. Epidemiological aspects and world distribution of HTLV-1 infection. Front Microbiol 3:388. doi: 10.3389/fmicb.2012.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filippone C, Betsem E, Tortevoye P, Cassar O, Bassot S, Froment A, Fontanet A, Gessain A. 2015. A severe bite from a nonhuman primate is a major risk factor for HTLV-1 infection in hunters from Central Africa. Clin Infect Dis 60:1667–1676. doi: 10.1093/cid/civ145. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe T. 2011. Current status of HTLV-1 infection. Int J Hematol 94:430–434. doi: 10.1007/s12185-011-0934-4. [DOI] [PubMed] [Google Scholar]
- 5.Qiu X, Hodges S, Lukaszewska T, Hino S, Arai H, Yamaguchi J, Swanson P, Schochetman G, Devare SG. 2008. Evaluation of a new, fully automated immunoassay for detection of HTLV-I and HTLV-II antibodies. J Med Virol 80:484–493. doi: 10.1002/jmv.21083. [DOI] [PubMed] [Google Scholar]
- 6.Cassar O, Gessain A. 2017. Serological and molecular methods to study epidemiological aspects Of Human T-cell lymphotropic virus type 1 infection. Methods Mol Biol 1582:3–24. doi: 10.1007/978-1-4939-6872-5_1. [DOI] [PubMed] [Google Scholar]
- 7.Miyakoshi H, Sugimoto M, Igarashi H, Honda H, Fujino R, Mizukoshi M. 1992. Improvement of simultaneous detection of antibodies to Gag and envelope antigens of human T-lymphotropic virus type I by Western immunoblot assay. J Clin Microbiol 30:2555–2559. http://jcm.asm.org/content/30/10/2555.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishizono I, Iida S, Suzuki N, Kawada H, Murakami H, Ashihara Y, Okada M. 1991. Rapid and sensitive chemiluminescent enzyme immunoassay for measuring tumor markers. Clin Chem 37:1639–1644. [PubMed] [Google Scholar]
- 9.Ikeda M, Fujino R, Matsui T, Yoshida T, Komoda H, Imai J. 1984. A new agglutination test for serum antibodies to adult T-cell leukemia virus. Gan 75:845–848. [PubMed] [Google Scholar]
- 10.Anonymous. 1991. AIDS: proposed WHO criteria for interpreting Western blot assays for HIV-1, HIV-2, and HTLV-I/HTLV-II. Bull World Health Organ 69:127–129, 131–133. [PMC free article] [PubMed] [Google Scholar]
- 11.Filippone C, Bassot S, Betsem E, Tortevoye P, Guillotte M, Mercereau-Puijalon O, Plancoulaine S, Calattini S, Gessain A. 2012. A new and frequent human T-cell leukemia virus indeterminate Western blot pattern: epidemiological determinants and PCR results in Central African inhabitants. J Clin Microbiol 50:1663–1672. doi: 10.1128/JCM.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garin B, Gosselin S, de The G, Gessain A. 1994. HTLV-I/II infection in a high viral endemic area of Zaire, Central Africa: comparative evaluation of serology, PCR, and significance of indeterminate Western blot pattern. J Med Virol 44:104–109. doi: 10.1002/jmv.1890440119. [DOI] [PubMed] [Google Scholar]
- 13.Porter KR, Liang L, Long GW, Bangs MJ, Anthony R, Andersen EM, Hayes CG. 1994. Evidence for anti-Plasmodium falciparum antibodies that cross-react with human T-lymphotropic virus type I proteins in a population in Irian Jaya, Indonesia. Clin Diagn Lab Immunol 1:11–15. http://cvi.asm.org/content/1/1/11.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahieux R, Horal P, Mauclere P, Mercereau-Puijalon O, Guillotte M, Meertens L, Murphy E, Gessain A. 2000. Human T-cell lymphotropic virus type 1 gag indeterminate Western blot patterns in Central Africa: relationship to Plasmodium falciparum infection. J Clin Microbiol 38:4049–4057. http://jcm.asm.org/content/38/11/4049.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes CG, Burans JP, Oberst RB. 1991. Antibodies to human T lymphotropic virus type I in a population from the Philippines: evidence for cross-reactivity with Plasmodium falciparum. J Infect Dis 163:257–262. doi: 10.1093/infdis/163.2.257. [DOI] [PubMed] [Google Scholar]
- 16.Calattini S, Betsem E, Bassot S, Chevalier SA, Mahieux R, Froment A, Gessain A. 2009. New strain of human T lymphotropic virus (HTLV) type 3 in a Pygmy from Cameroon with peculiar HTLV serologic results. J Infect Dis 199:561–564. doi: 10.1086/596206. [DOI] [PubMed] [Google Scholar]
- 17.Calattini S, Chevalier SA, Duprez R, Bassot S, Froment A, Mahieux R, Gessain A. 2005. Discovery of a new human T-cell lymphotropic virus (HTLV-3) in Central Africa. Retrovirology 2:30. doi: 10.1186/1742-4690-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martins ML, Santos AC, Namen-Lopes MS, Barbosa-Stancioli EF, Utsch DG, Carneiro-Proietti AB. 2010. Long-term serological follow-up of blood donors with an HTLV-indeterminate Western blot: antibody profile of seroconverters and individuals with false reactions. J Med Virol 82:1746–1753. doi: 10.1002/jmv.21881. [DOI] [PubMed] [Google Scholar]
- 19.Yao K, Hisada M, Maloney E, Yamano Y, Hanchard B, Wilks R, Rios M, Jacobson S. 2006. Human T lymphotropic virus types I and II Western blot seroindeterminate status and its association with exposure to prototype HTLV-I. J Infect Dis 193:427–437. doi: 10.1086/499273. [DOI] [PubMed] [Google Scholar]
- 20.Okayama A, Stuver S, Iga M, Okamoto M, Mueller N, Matsuoka M, Yamaguchi K, Tachibana N, Tsubouchi H. 2001. Sequential change of virus markers in seroconverters with community-acquired infection of human T lymphotropic virus type I. J Infect Dis 183:1031–1037. doi: 10.1086/319282. [DOI] [PubMed] [Google Scholar]
- 21.Césaire R, Bera O, Maier H, Lezin A, Martial J, Ouka M, Kerob-Bauchet B, Ould Amar AK, Vernant JC. 1999. Seroindeterminate patterns and seroconversions to human T-lymphotropic virus type I positivity in blood donors from Martinique, French West Indies. Transfusion 39:1145–1149. doi: 10.1046/j.1537-2995.1999.39101145.x. [DOI] [PubMed] [Google Scholar]
- 22.Manns A, Murphy EL, Wilks R, Haynes G, Figueroa JP, Hanchard B, Barnett M, Drummond J, Waters D, Cerney M. 1991. Detection of early human T-cell lymphotropic virus type I antibody patterns during seroconversion among transfusion recipients. Blood 77:896–905. [PubMed] [Google Scholar]
- 23.Rouet F, Meertens L, Courouble G, Herrmann-Storck C, Pabingui R, Chancerel B, Abid A, Strobel M, Mauclere P, Gessain A. 2001. Serological, epidemiological, and molecular differences between human T-cell lymphotropic virus type 1 (HTLV-1)-seropositive healthy carriers and persons with HTLV-I Gag indeterminate Western blot patterns from the Caribbean. J Clin Microbiol 39:1247–1253. doi: 10.1128/JCM.39.4.1247-1253.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanjani DS, Shahabi M, Talaei N, Afzalaghaee M, Tehranian F, Bazargani R. 2011. Molecular analysis of human T cell lymphotropic virus type 1 and 2 (HTLV-1/2) seroindeterminate blood donors from northeast Iran: evidence of proviral tax, env, and gag sequences. AIDS Res Hum Retroviruses 27:131–135. doi: 10.1089/aid.2010.0017. [DOI] [PubMed] [Google Scholar]
- 25.Berini CA, Eirin ME, Pando MA, Biglione MM. 2007. Human T-cell lymphotropic virus types I and II (HTLV-I and -II) infection among seroindeterminate cases in Argentina. J Med Virol 79:69–73. doi: 10.1002/jmv.20731. [DOI] [PubMed] [Google Scholar]
- 26.Costa JM, Segurado AC. 2009. Molecular evidence of human T-cell lymphotropic virus types 1 and 2 (HTLV-1 and HTLV-2) infections in HTLV seroindeterminate individuals from Sao Paulo, Brazil. J Clin Virol 44:185–189. doi: 10.1016/j.jcv.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 27.Mangano AM, Remesar M, del Pozo A, Sen L. 2004. Human T lymphotropic virus types I and II proviral sequences in Argentinian blood donors with indeterminate Western blot patterns. J Med Virol 74:323–327. doi: 10.1002/jmv.20172. [DOI] [PubMed] [Google Scholar]
- 28.Matsumoto C, Sagara Y, Sobata R, Inoue Y, Morita M, Uchida S, Kiyokawa H, Satake M, Tadokoro K. 2017. Analysis of HTLV-1 proviral load (PVL) and antibody detected with various kinds of tests in Japanese blood donors to understand the relationship between PVL and antibody level and to gain insights toward better antibody testing. J Med Virol 89:1469–1476. doi: 10.1002/jmv.24802. [DOI] [PubMed] [Google Scholar]
- 29.Fan J, Ma G, Nosaka K, Tanabe J, Satou Y, Koito A, Wain-Hobson S, Vartanian JP, Matsuoka M. 2010. APOBEC3G generates nonsense mutations in human T-cell leukemia virus type 1 proviral genomes in vivo. J Virol 84:7278–7287. doi: 10.1128/JVI.02239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georgoulias VA, Malliaraki NE, Theodoropoulou M, Spanakis E, Fountouli P, Tsatsaki D, Kotsaki S, Karvela-Aggelaki A, Malliaraki-Pinetidou E. 1997. Indeterminate human immunodeficiency virus type 1 Western blot may indicate an abortive infection in some low-risk blood donors. Transfusion 37:65–72. doi: 10.1046/j.1537-2995.1997.37197176953.x. [DOI] [PubMed] [Google Scholar]
- 31.Cánepa C, Salido J, Ruggieri M, Fraile S, Pataccini G, Berini C, Biglione M. 2015. Low proviral load is associated with indeterminate Western blot patterns in human T-cell lymphotropic virus type 1 infected individuals: could punctual mutations be related? Viruses 7:5643–5658. doi: 10.3390/v7112897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith MR, Greene WC. 1990. Identification of HTLV-I tax trans-activator mutants exhibiting novel transcriptional phenotypes. Genes Dev 4:1875–1885. doi: 10.1101/gad.4.11.1875. [DOI] [PubMed] [Google Scholar]
- 33.Younis I, Green PL. 2005. The human T-cell leukemia virus Rex protein. Front Biosci 10:431–445. doi: 10.2741/1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, St John A, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med 10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 35.Friedrich TC, Dodds EJ, Yant LJ, Vojnov L, Rudersdorf R, Cullen C, Evans DT, Desrosiers RC, Mothe BR, Sidney J, Sette A, Kunstman K, Wolinsky S, Piatak M, Lifson J, Hughes AL, Wilson N, O'Connor DH, Watkins DI. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat Med 10:275–281. doi: 10.1038/nm998. [DOI] [PubMed] [Google Scholar]
- 36.Carpentier A, Barez PY, Hamaidia M, Gazon H, de Brogniez A, Perike S, Gillet N, Willems L. 2015. Modes of human T cell leukemia virus type 1 transmission, replication and persistence. Viruses 7:3603–3624. doi: 10.3390/v7072793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol 11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 38.Sugamura K, Fujii M, Kannagi M, Sakitani M, Takeuchi M, Hinuma Y. 1984. Cell surface phenotypes and expression of viral antigens of various human cell lines carrying human T-cell leukemia virus. Int J Cancer 34:221–228. doi: 10.1002/ijc.2910340213. [DOI] [PubMed] [Google Scholar]
- 39.Kuramitsu M, Okuma K, Yamagishi M, Yamochi T, Firouzi S, Momose H, Mizukami T, Takizawa K, Araki K, Sugamura K, Yamaguchi K, Watanabe T, Hamaguchi I. 2015. Identification of TL-Om1, an adult T-cell leukemia (ATL) cell line, as reference material for quantitative PCR for human T-lymphotropic virus 1. J Clin Microbiol 53:587–596. doi: 10.1128/JCM.02254-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuramitsu M, Okuma K, Yamochi T, Sato T, Sasaki D, Hasegawa H, Umeki K, Kubota R, Sobata R, Matsumoto C, Kaneko N, Naruse I, Yamagishi M, Nakashima M, Momose H, Araki K, Mizukami T, Mizusawa S, Okada Y, Ochiai M, Utsunomiya A, Koh KR, Ogata M, Nosaka K, Uchimaru K, Iwanaga M, Sagara Y, Yamano Y, Satake M, Okayama A, Mochizuki M, Izumo S, Saito S, Itabashi K, Kamihira S, Yamaguchi K, Watanabe T, Hamaguchi I. 2015. Standardization of quantitative PCR for human T-cell leukemia virus type 1 in Japan: a collaborative study. J Clin Microbiol 53:3485–3491. doi: 10.1128/JCM.01628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sobata R, Matsumoto C, Uchida S, Suzuki Y, Satake M, Tadokoro K. 2015. Estimation of the infectious viral load required for transfusion-transmitted human T-lymphotropic virus type 1 infection (TT-HTLV-1) and of the effectiveness of leukocyte reduction in preventing TT-HTLV-1. Vox Sang 109:122–128. doi: 10.1111/vox.12263. [DOI] [PubMed] [Google Scholar]
- 42.Ueno S, Umeki K, Takajo I, Nagatomo Y, Kusumoto N, Umekita K, Morishita K, Okayama A. 2012. Proviral loads of human T-lymphotropic virus type 1 in asymptomatic carriers with different infection routes. Int J Cancer 130:2318–2326. doi: 10.1002/ijc.26289. [DOI] [PubMed] [Google Scholar]
- 43.Miyazato P, Yasunaga J, Taniguchi Y, Koyanagi Y, Mitsuya H, Matsuoka M. 2006. De novo human T-cell leukemia virus type 1 infection of human lymphocytes in NOD-SCID, common gamma-chain knockout mice. J Virol 80:10683–10691. doi: 10.1128/JVI.01009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe M, Ohsugi T, Shoda M, Ishida T, Aizawa S, Maruyama-Nagai M, Utsunomiya A, Koga S, Yamada Y, Kamihira S, Okayama A, Kikuchi H, Uozumi K, Yamaguchi K, Higashihara M, Umezawa K, Watanabe T, Horie R. 2005. Dual targeting of transformed and untransformed HTLV-1-infected T cells by DHMEQ, a potent and selective inhibitor of NF-kappaB, as a strategy for chemoprevention and therapy of adult T-cell leukemia. Blood 106:2462–2471. doi: 10.1182/blood-2004-09-3646. [DOI] [PubMed] [Google Scholar]
- 45.Takenouchi N, Yamano Y, Usuku K, Osame M, Izumo S. 2003. Usefulness of proviral load measurement for monitoring of disease activity in individual patients with human T-lymphotropic virus type I-associated myelopathy/tropical spastic paraparesis. J Neurovirol 9:29–35. doi: 10.1080/13550280390173418. [DOI] [PubMed] [Google Scholar]
- 46.Kamihira S, Dateki N, Sugahara K, Hayashi T, Harasawa H, Minami S, Hirakata Y, Yamada Y. 2003. Significance of HTLV-1 proviral load quantification by real-time PCR as a surrogate marker for HTLV-1-infected cell count. Clin Lab Haematol 25:111–117. doi: 10.1046/j.1365-2257.2003.00503.x. [DOI] [PubMed] [Google Scholar]
- 47.Nagai M, Yamano Y, Brennan MB, Mora CA, Jacobson S. 2001. Increased HTLV-I proviral load and preferential expansion of HTLV-I Tax-specific CD8+ T cells in cerebrospinal fluid from patients with HAM/TSP. Ann Neurol 50:807–812. doi: 10.1002/ana.10065. [DOI] [PubMed] [Google Scholar]
- 48.Cassar O, Einsiedel L, Afonso PV, Gessain A. 2013. Human T-cell lymphotropic virus type 1 subtype C molecular variants among indigenous Australians: new insights into the molecular epidemiology of HTLV-1 in Australo-Melanesia. PLoS Negl Trop Dis 7:e2418. doi: 10.1371/journal.pntd.0002418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.